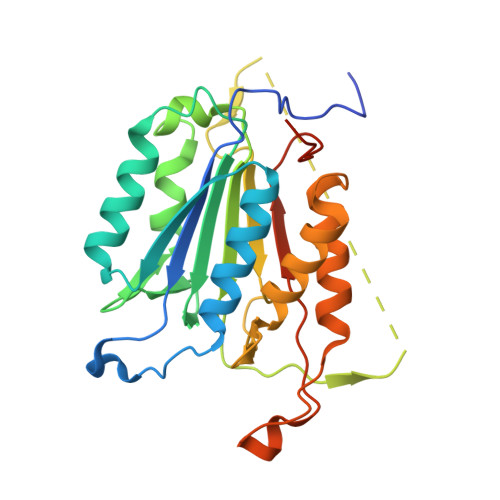
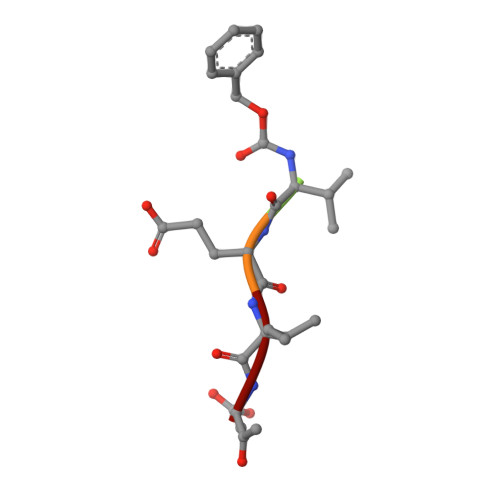
Mechanistic and structural understanding of uncompetitive inhibitors of caspase-6.
Heise, C.E., Murray, J., Augustyn, K.E., Bravo, B., Chugha, P., Cohen, F., Giannetti, A.M., Gibbons, P., Hannoush, R.N., Hearn, B.R., Jaishankar, P., Ly, C.Q., Shah, K., Stanger, K., Steffek, M., Tang, Y., Zhao, X., Lewcock, J.W., Renslo, A.R., Flygare, J., Arkin, M.R.(2012) PLoS One 7: e50864-e50864
- PubMed: 23227217
- DOI: https://doi.org/10.1371/journal.pone.0050864
- Primary Citation of Related Structures:
4HVA - PubMed Abstract:
Inhibition of caspase-6 is a potential therapeutic strategy for some neurodegenerative diseases, but it has been difficult to develop selective inhibitors against caspases. We report the discovery and characterization of a potent inhibitor of caspase-6 that acts by an uncompetitive binding mode that is an unprecedented mechanism of inhibition against this target class. Biochemical assays demonstrate that, while exquisitely selective for caspase-6 over caspase-3 and -7, the compound's inhibitory activity is also dependent on the amino acid sequence and P1' character of the peptide substrate. The crystal structure of the ternary complex of caspase-6, substrate-mimetic and an 11 nM inhibitor reveals the molecular basis of inhibition. The general strategy to develop uncompetitive inhibitors together with the unique mechanism described herein provides a rationale for engineering caspase selectivity.
- Department of Biochemical and Cellular Pharmacology, Genentech, Inc, South San Francisco, California, United States of America. heise.christopher@gene.com
Organizational Affiliation: