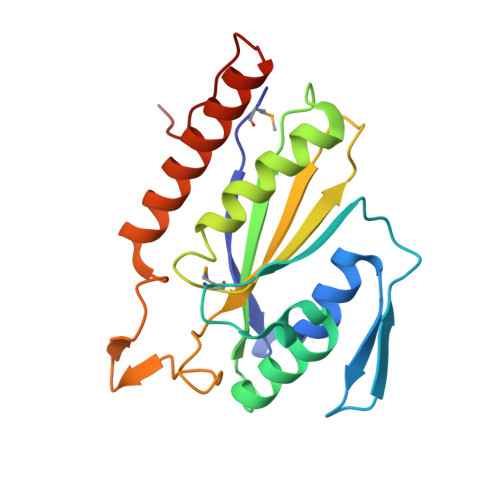
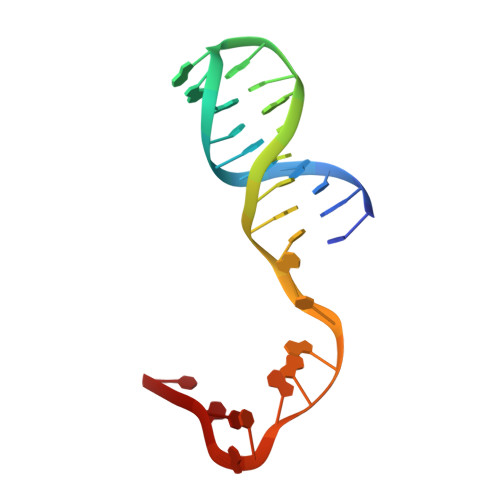
Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus.
Edwards, J.S., Betts, L., Frazier, M.L., Pollet, R.M., Kwong, S.M., Walton, W.G., Ballentine, W.K., Huang, J.J., Habibi, S., Del Campo, M., Meier, J.L., Dervan, P.B., Firth, N., Redinbo, M.R.(2013) Proc Natl Acad Sci U S A 110: 2804-2809
- PubMed: 23359708
- DOI: https://doi.org/10.1073/pnas.1219701110
- Primary Citation of Related Structures:
4HT4, 4HTE - PubMed Abstract:
Multidrug-resistant Staphylococcus aureus infections pose a significant threat to human health. Antibiotic resistance is most commonly propagated by conjugative plasmids like pLW1043, the first vancomycin-resistant S. aureus vector identified in humans. We present the molecular basis for resistance transmission by the nicking enzyme in S. aureus (NES), which is essential for conjugative transfer. NES initiates and terminates the transfer of plasmids that variously confer resistance to a range of drugs, including vancomycin, gentamicin, and mupirocin. The NES N-terminal relaxase-DNA complex crystal structure reveals unique protein-DNA contacts essential in vitro and for conjugation in S. aureus. Using this structural information, we designed a DNA minor groove-targeted polyamide that inhibits NES with low micromolar efficacy. The crystal structure of the 341-residue C-terminal region outlines a unique architecture; in vitro and cell-based studies further establish that it is essential for conjugation and regulates the activity of the N-terminal relaxase. This conclusion is supported by a small-angle X-ray scattering structure of a full-length, 665-residue NES-DNA complex. Together, these data reveal the structural basis for antibiotic multiresistance acquisition by S. aureus and suggest novel strategies for therapeutic intervention.
- Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: