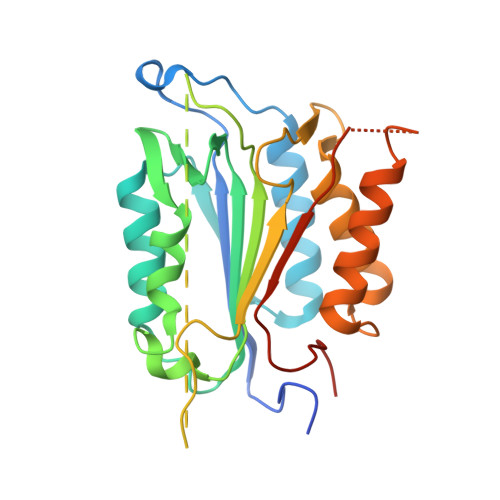
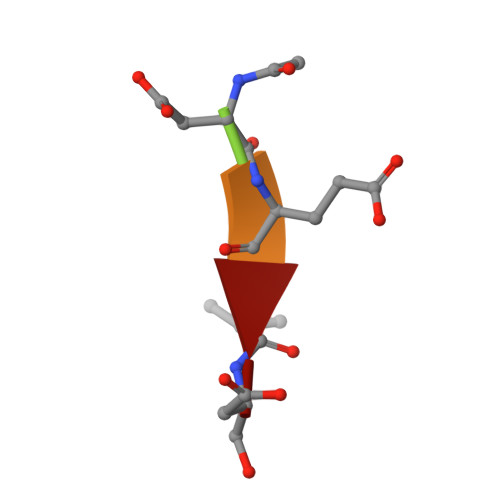
Structural asymmetry of procaspase-7 bound to a specific inhibitor
Kang, H.J., Lee, Y.M., Bae, K.H., Kim, S.J., Chung, S.J.(2013) Acta Crystallogr D Biol Crystallogr 69: 1514-1521
- PubMed: 23897474
- DOI: https://doi.org/10.1107/S0907444913010196
- Primary Citation of Related Structures:
4HQ0, 4HQR - PubMed Abstract:
Caspase-7 is expressed as a proenzyme and is activated by initiator caspases upon the transmission of cell-death signals. Despite extensive structural and biochemical analyses, many questions regarding the mechanism of caspase-7 activation remain unanswered. Caspase-7 is auto-activated during overexpression in Escherichia coli, even in the absence of initiator caspases, indicating that procaspase-7 has intrinsic catalytic activity. When variants of procaspase-7 with altered L2 loops were prepared, a variant with six inserted amino acids showed meaningful catalytic activity which was inhibited by Ac-DEVD-CHO. The kinetic constants of the procaspase-7 variant were determined and its three-dimensional structure was solved with and without bound inhibitor. The homodimeric procaspase-7 bound to the inhibitor revealed an asymmetry. One monomer formed a complete active site bound to the inhibitor in collaboration with the L2 loop from the other monomer, whereas the other monomer had an incomplete active site despite the bound inhibitor. Consequently, the two L2 loops in homodimeric procaspase-7 served as inherent L2 and L2' loops forming one complete active site. These data represent the first three-dimensional structure of a procaspase-7 variant bound to a specific inhibitor, Ac-DEVD-CHO, and provide insight into the folding mechanism during caspase-7 activation and the basal activity level of procaspase-7.
- Department of Chemistry, College of Natural Science, Dongguk University, 26 Pil-dong 3-ga, Jung-gu, Seoul 100-715, Republic of Korea.
Organizational Affiliation: