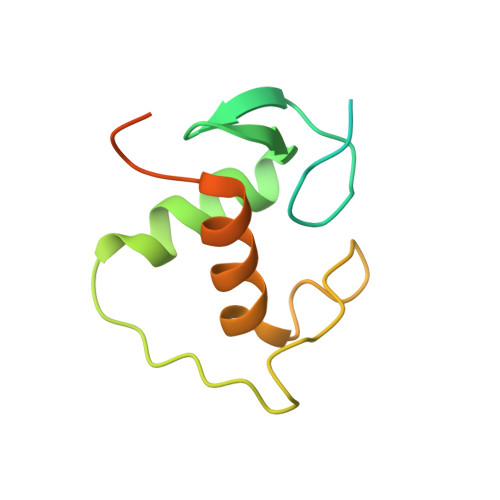
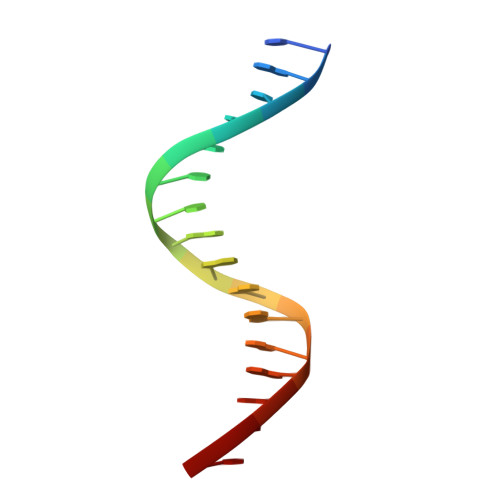
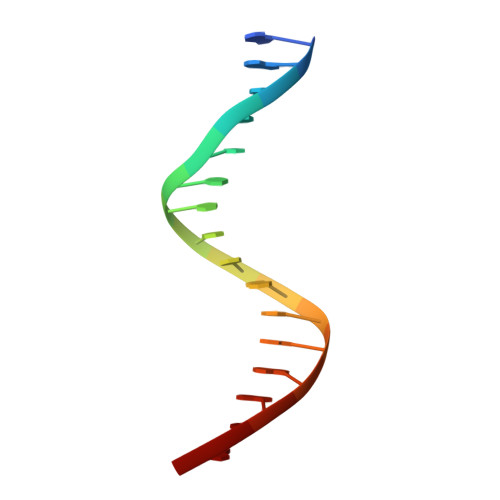
The structural basis of direct glucocorticoid-mediated transrepression.
Hudson, W.H., Youn, C., Ortlund, E.A.(2013) Nat Struct Mol Biol 20: 53-58
- PubMed: 23222642
- DOI: https://doi.org/10.1038/nsmb.2456
- Primary Citation of Related Structures:
4HN5, 4HN6 - PubMed Abstract:
A newly discovered negative glucocorticoid response element (nGRE) mediates DNA-dependent transrepression by the glucocorticoid receptor (GR) across the genome and has a major role in immunosuppressive therapy. The nGRE differs dramatically from activating response elements, and the mechanism driving GR binding and transrepression is unknown. To unravel the mechanism of nGRE-mediated transrepression by the GR, we characterized the interaction between GR and an nGRE in the thymic stromal lymphopoietin (TSLP) promoter. We show using structural and mechanistic approaches that nGRE binding is a new mode of sequence recognition by human GR and that nGREs prevent receptor dimerization through a unique GR-binding orientation and strong negative cooperativity, ensuring the presence of monomeric GR at repressive elements.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia, USA.
Organizational Affiliation: