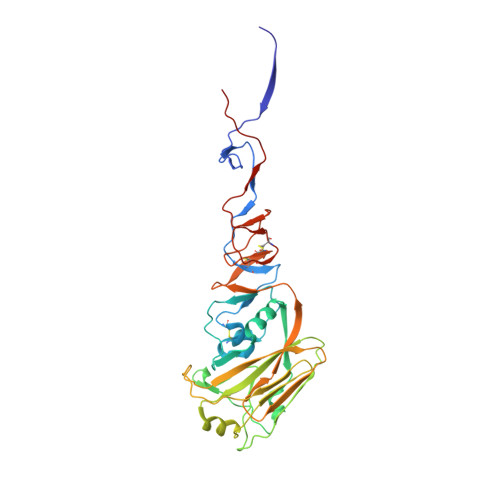
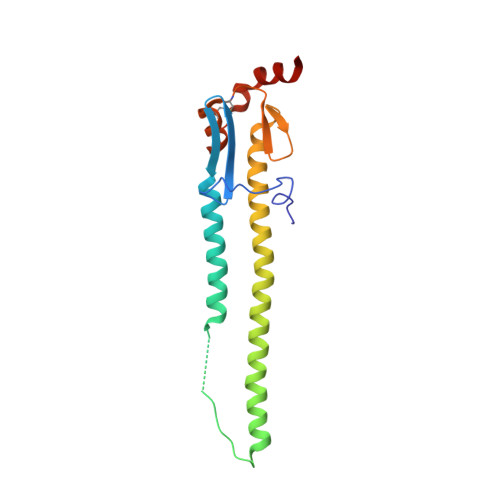
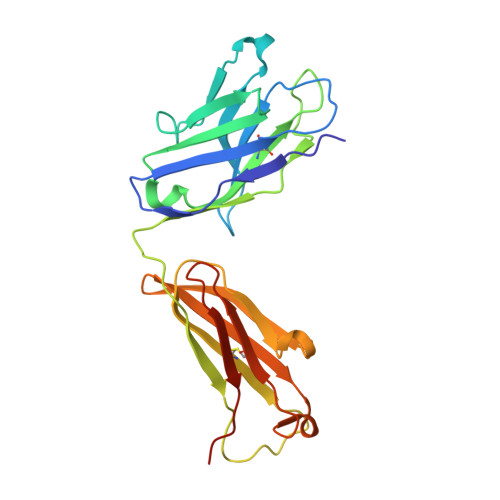
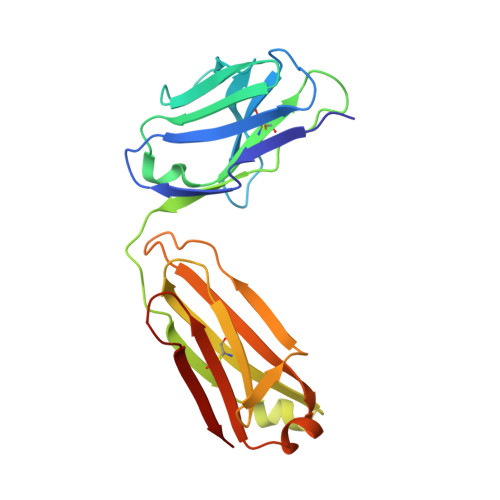
Structure of a classical broadly neutralizing stem antibody in complex with a pandemic h2 influenza virus hemagglutinin.
Dreyfus, C., Ekiert, D.C., Wilson, I.A.(2013) J Virol 87: 7149-7154
- PubMed: 23552413
- DOI: https://doi.org/10.1128/JVI.02975-12
- Primary Citation of Related Structures:
4HLZ - PubMed Abstract:
We report the structural characterization of the first antibody identified to cross-neutralize multiple subtypes of influenza A viruses. The crystal structure of mouse antibody C179 bound to the pandemic 1957 H2N2 hemagglutinin (HA) reveals that it targets an epitope on the HA stem similar to those targeted by the recently identified human broadly neutralizing antibodies. C179 also inhibits the low-pH conformational change of the HA but uses a different angle of approach and both heavy and light chains.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, USA.
Organizational Affiliation: