Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies.
Donaldson, J.M., Zer, C., Avery, K.N., Bzymek, K.P., Horne, D.A., Williams, J.C.(2013) Proc Natl Acad Sci U S A 110: 17456-17461
- PubMed: 24101516
- DOI: https://doi.org/10.1073/pnas.1307309110
- Primary Citation of Related Structures:
4GW1, 4GW5, 4HJG, 4HKZ, 4IOI - PubMed Abstract:
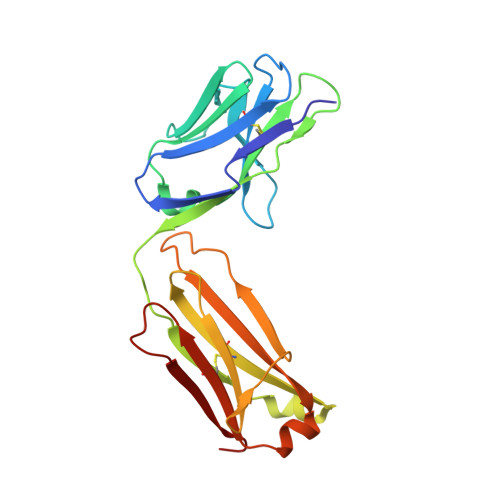
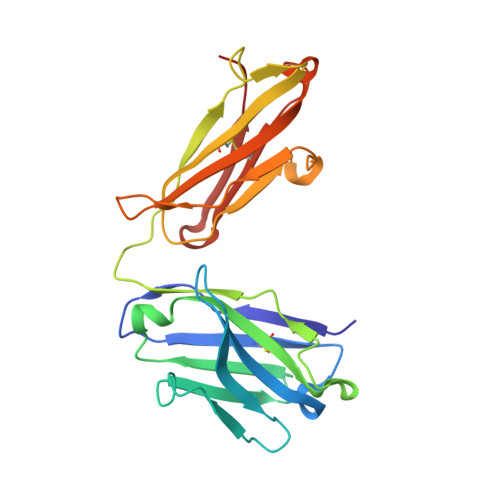
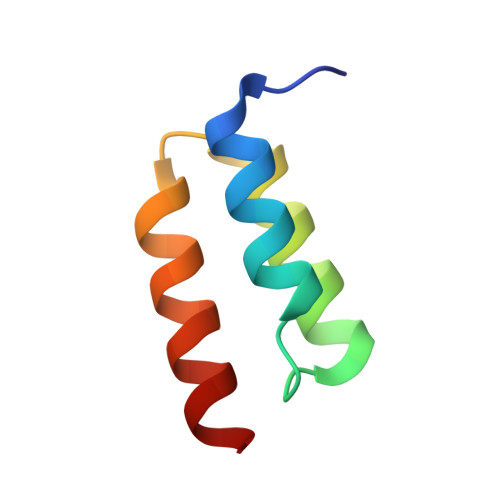
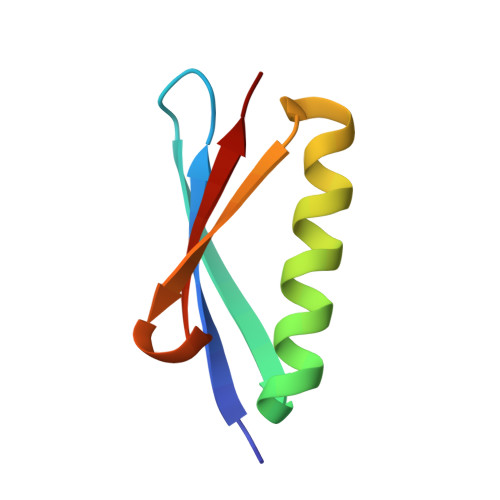
Capitalizing on their extraordinary specificity, monoclonal antibodies (mAbs) have become one of the most reengineered classes of biological molecules. A major goal in many of these engineering efforts is to add new functionality to the parental mAb, including the addition of cytotoxins and imaging agents for medical applications. Herein, we present a unique peptide-binding site within the central cavity of the fragment antigen binding framework region of the chimeric, anti-epidermal growth factor receptor mAb cetuximab. We demonstrate through diffraction methods, biophysical studies, and sequence analysis that this peptide, a meditope, has moderate affinity for the Fab, is specific to cetuximab (i.e., does not bind to human IgGs), and has no significant effect on antigen binding. We further demonstrate by diffraction studies and biophysical methods that the meditope binding site can be grafted onto the anti-human epidermal growth factor receptor 2 mAb trastuzumab, and that the antigen binding affinity of the grafted trastuzumab is indistinguishable from the parental mAb. Finally, we demonstrate a bivalent meditope variant binds specifically and stably to antigen-bearing cells only in the presence of the meditope-enabled mAbs. Collectively, this finding and the subsequent characterization and engineering efforts indicate that this unique interface could serve as a noncovalent "linker" for any meditope-enabled mAb with applications in multiple mAb-based technologies including diagnostics, imaging, and therapeutic delivery.
- Department of Molecular Medicine, Beckman Research Institute of City of Hope, Duarte, CA 91010.
Organizational Affiliation: