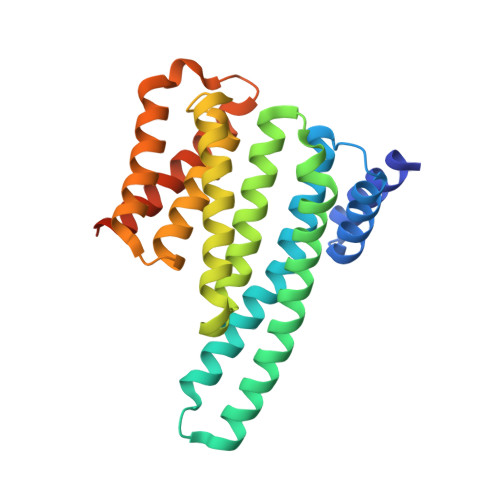
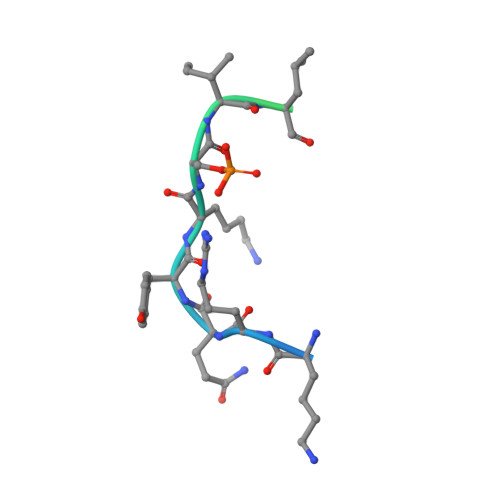
Characterization of 14-3-3-zeta Interactions with integrin tails
Bonet, R., Vakonakis, I., Campbell, I.D.(2013) J Mol Biology 425: 3060-3072
- PubMed: 23763993
- DOI: https://doi.org/10.1016/j.jmb.2013.05.024
- Primary Citation of Related Structures:
4HKC - PubMed Abstract:
Integrins are a family of heterodimeric (α+β) adhesion receptors that play key roles in many cellular processes. Integrins are unusual in that their functions can be modulated from both outside and inside the cell. Inside-out signaling is mediated by binding adaptor proteins to the flexible cytoplasmic tails of the α- and β-integrin subunits. Talin is one well-known intracellular activator, but various other adaptors bind to integrin tails, including 14-3-3-ζ, a member of the 14-3-3 family of dimeric proteins that have a preference for binding phosphorylated sequence motifs. Phosphorylation of a threonine in the β2 integrin tail has been shown to modulate β2/14-3-3-ζ interactions, and recently, the α4 integrin tail was reported to bind to 14-3-3-ζ and associate with paxillin in a ternary complex that is regulated by serine phosphorylation. Here, we use a range of biophysical techniques to characterize interactions between 14-3-3-ζ and the cytoplasmic tails of α4, β1, β2 and β3 integrins. The X-ray structure of the 14-3-3-ζ/α4 complex indicates a canonical binding mode for the α4 phospho-peptide, but unexpected features are also observed: residues outside the consensus 14-3-3-ζ binding motif are shown to be essential for an efficient interaction; in contrast, a short β2 phospho-peptide is sufficient for high-affinity binding to 14-3-3-ζ. In addition, we report novel 14-3-3-ζ/integrin tail interactions that are independent of phosphorylation. Of the integrin tails studied, the strongest interaction with 14-3-3-ζ is observed for the β1A variant. In summary, new insights about 14-3-3-ζ/integrin tail interactions that have implications for the role of these molecular associations in cells are described.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, United Kingdom. roman.bonet@iqac.csic.es
Organizational Affiliation: