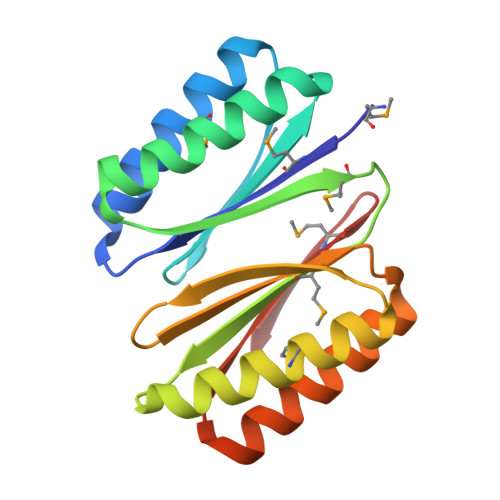
Crystal Structure of Engineered Protein OR280.
Vorobiev, S., Lew, S., Lin, Y.-R., Seetharaman, J., Castelllanos, J., Maglaqui, M., Xiao, R., Lee, D., Koga, N., Koga, R., Everett, J.K., Acton, T.B., Baker, D., Montelione, G.T., Tong, L., Hunt, J.F.To be published.