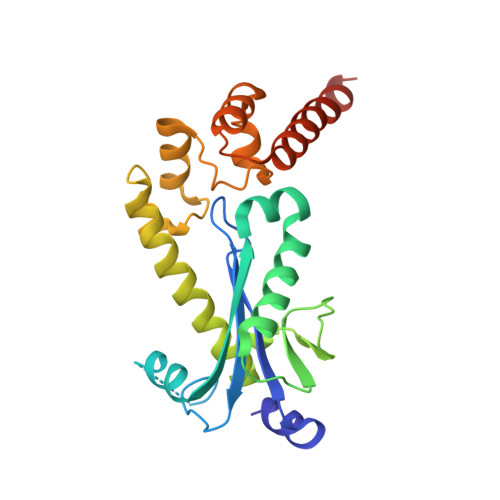
RNase H2 roles in genome integrity revealed by unlinking its activities.
Chon, H., Sparks, J.L., Rychlik, M., Nowotny, M., Burgers, P.M., Crouch, R.J., Cerritelli, S.M.(2013) Nucleic Acids Res 41: 3130-3143
- PubMed: 23355612
- DOI: https://doi.org/10.1093/nar/gkt027
- Primary Citation of Related Structures:
4HHT - PubMed Abstract:
Ribonuclease H2 (RNase H2) protects genome integrity by its dual roles of resolving transcription-related R-loops and ribonucleotides incorporated in DNA during replication. To unlink these two functions, we generated a Saccharomyces cerevisiae RNase H2 mutant that can resolve R-loops but cannot cleave single ribonucleotides in DNA. This mutant definitively correlates the 2-5 bp deletions observed in rnh201Δ strains with single rNMPs in DNA. It also establishes a connection between R-loops and Sgs1-mediated replication reinitiation at stalled forks and identifies R-loops uniquely processed by RNase H2. In mouse, deletion of any of the genes coding for RNase H2 results in embryonic lethality, and in humans, RNase H2 hypomorphic mutations cause Aicardi-Goutières syndrome (AGS), a neuroinflammatory disorder. To determine the contribution of R-loops and rNMP in DNA to the defects observed in AGS, we characterized in yeast an AGS-related mutation, which is impaired in processing both substrates, but has sufficient R-loop degradation activity to complement the defects of rnh201Δ sgs1Δ strains. However, this AGS-related mutation accumulates 2-5 bp deletions at a very similar rate as the deletion strain.
- Program in Genomics of Differentiation, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD 20892, USA.
Organizational Affiliation: