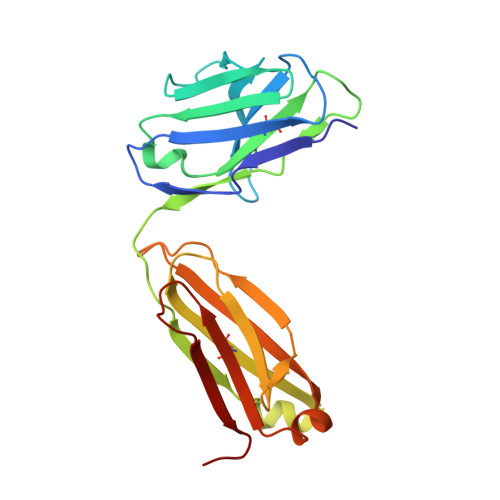
Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
Bryson, S., Thomson, C.A., Risnes, L.F., Dasgupta, S., Smith, K., Schrader, J.W., Pai, E.F.(2016) J Immunol 196: 4723-4730
- PubMed: 27183571
- DOI: https://doi.org/10.4049/jimmunol.1402890
- Primary Citation of Related Structures:
4HH9, 4HHA, 4HIE, 4HIH, 4HII, 4HIJ, 4PTT, 4PTU - PubMed Abstract:
The human Ab response to certain pathogens is oligoclonal, with preferred IgV genes being used more frequently than others. A pair of such preferred genes, IGVK3-11 and IGVH3-30, contributes to the generation of protective Abs directed against the 23F serotype of the pneumonococcal capsular polysaccharide of Streptococcus pneumoniae and against the AD-2S1 peptide of the gB membrane protein of human CMV. Structural analyses of Fab fragments of mAbs 023.102 and pn132p2C05 in complex with portions of the 23F polysaccharide revealed five germline-encoded residues in contact with the key component, l-rhamnose. In the case of the AD-2S1 peptide, the KE5 Fab fragment complex identified nine germline-encoded contact residues. Two of these germline-encoded residues, Arg91L and Trp94L, contact both the l-rhamnose and the AD-2S1 peptide. Comparison of the respective paratopes that bind to carbohydrate and protein reveals that stochastic diversity in both CDR3 loops alone almost exclusively accounts for their divergent specificity. Combined evolutionary pressure by human CMV and the 23F serotype of S. pneumoniae acted on the IGVK3-11 and IGVH3-30 genes as demonstrated by the multiple germline-encoded amino acids that contact both l-rhamnose and AD-2S1 peptide.
- Princess Margaret Cancer Centre, Toronto, Ontario M5G 1L7, Canada; Department of Biochemistry, University of Toronto, Toronto, Ontario M5S 1A8, Canada;
Organizational Affiliation: