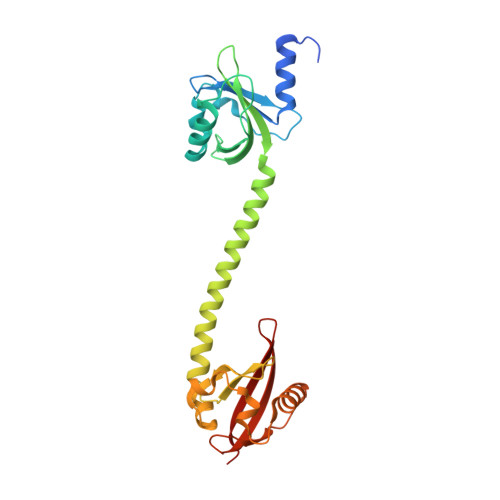
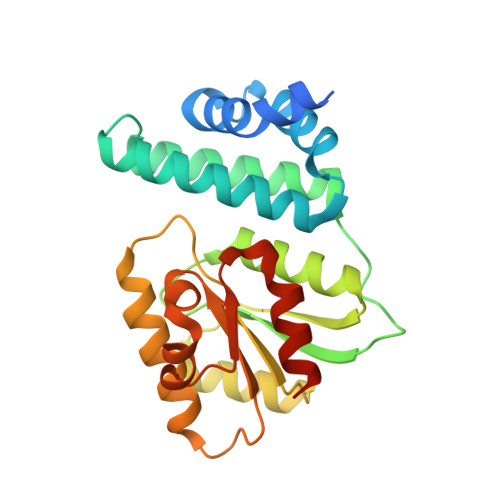
A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Winkler, A., Heintz, U., Lindner, R., Reinstein, J., Shoeman, R.L., Schlichting, I.(2013) Nat Struct Mol Biol 20: 859-867
- PubMed: 23728293
- DOI: https://doi.org/10.1038/nsmb.2597
- Primary Citation of Related Structures:
4HH0, 4HH1, 4HH2, 4HH3 - PubMed Abstract:
The anoxygenic phototrophic bacterium Rhodobacter sphaeroides uses different energy sources, depending on environmental conditions including aerobic respiration or, in the absence of oxygen, photosynthesis. Photosynthetic genes are repressed at high oxygen tension, but at intermediate levels their partial expression prepares the bacterium for using light energy. Illumination, however, enhances repression under semiaerobic conditions. Here, we describe molecular details of two proteins mediating oxygen and light control of photosynthesis-gene expression: the light-sensing antirepressor AppA and the transcriptional repressor PpsR. Our crystal structures of both proteins and their complex and hydrogen/deuterium-exchange data show that light activation of AppA-PpsR2 affects the PpsR effector region within the complex. DNA binding studies demonstrate the formation of a light-sensitive ternary AppA-PpsR-DNA complex. We discuss implications of these results for regulation by light and oxygen, highlighting new insights into blue light-mediated signal transduction.
- Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Heidelberg, Germany. andreas.winkler@mpimf-heidelberg.mpg.de
Organizational Affiliation: