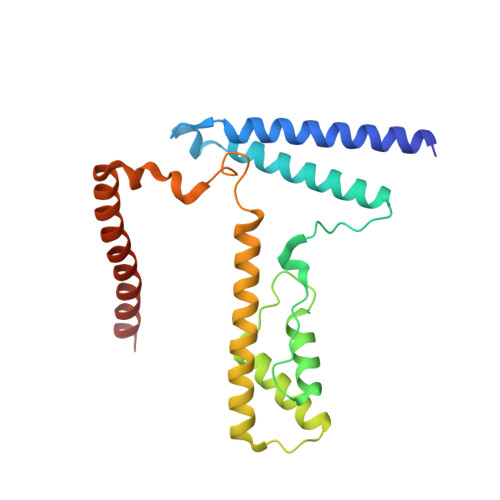
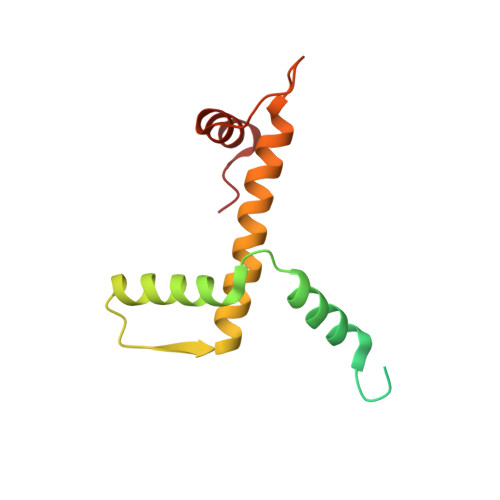
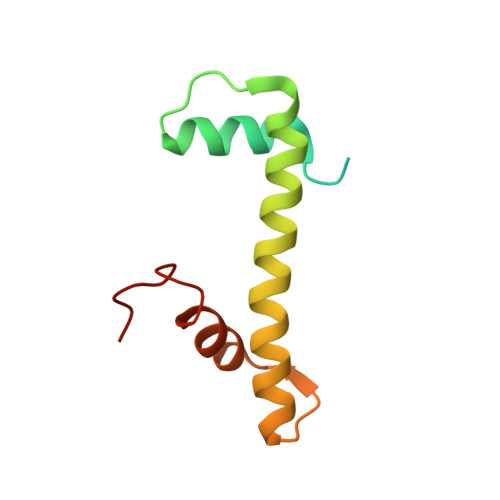
Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Liu, C.P., Xiong, C.Y., Wang, M.Z., Yu, Z.L., Yang, N., Chen, P., Zhang, Z.G., Li, G.H., Xu, R.M.(2012) Nat Struct Mol Biol 19: 1287-1292
- PubMed: 23142979
- DOI: https://doi.org/10.1038/nsmb.2439
- Primary Citation of Related Structures:
4HGA - PubMed Abstract:
Mammalian histone H3.3 is a variant of the canonical H3.1 essential for genome reprogramming in fertilized eggs and maintenance of chromatin structure in neuronal cells. An H3.3-specific histone chaperone, DAXX, directs the deposition of H3.3 onto pericentric and telomeric heterochromatin. H3.3 differs from H3.1 by only five amino acids, yet DAXX can distinguish the two with high precision. By a combination of structural, biochemical and cell-based targeting analyses, we show that Ala87 and Gly90 are the principal determinants of human H3.3 specificity. DAXX uses a shallow hydrophobic pocket to accommodate the small hydrophobic Ala87 of H3.3, whereas a polar binding environment in DAXX prefers Gly90 in H3.3 over the hydrophobic Met90 in H3.1. An H3.3-H4 heterodimer is bound by the histone-binding domain of DAXX, which makes extensive contacts with both H3.3 and H4.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: