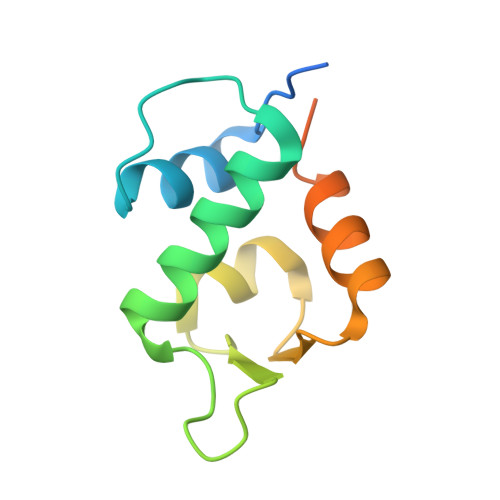
The structure of an MDM2-Nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant.
Anil, B., Riedinger, C., Endicott, J.A., Noble, M.E.(2013) Acta Crystallogr D Biol Crystallogr 69: 1358-1366
- PubMed: 23897459
- DOI: https://doi.org/10.1107/S0907444913004459
- Primary Citation of Related Structures:
4HFZ, 4HG7 - PubMed Abstract:
The p53-binding site of MDM2 holds great promise as a target for therapeutic intervention in MDM2-amplified p53 wild-type forms of cancer. Despite the extensive validation of this strategy, there are relatively few crystallographically determined co-complex structures for small-molecular inhibitors of the MDM2-p53 interaction available in the PDB. Here, a surface-entropy reduction mutant of the N-terminal domain of MDM2 that has been designed to enhance crystallogenesis is presented. This mutant has been validated by comparative ligand-binding studies using differential scanning fluorimetry and fluorescence polarization anisotropy and by cocrystallization with a peptide derived from p53. Using this mutant, the cocrystal structure of MDM2 with the benchmark inhibitor Nutlin-3a has been determined, revealing subtle differences from the previously described co-complex of MDM2 with Nutlin-2.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, England.
Organizational Affiliation: