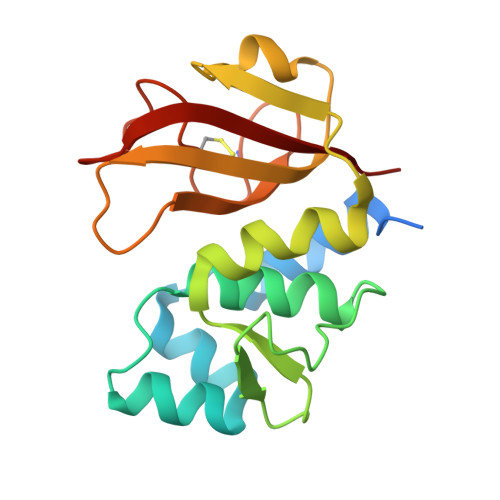
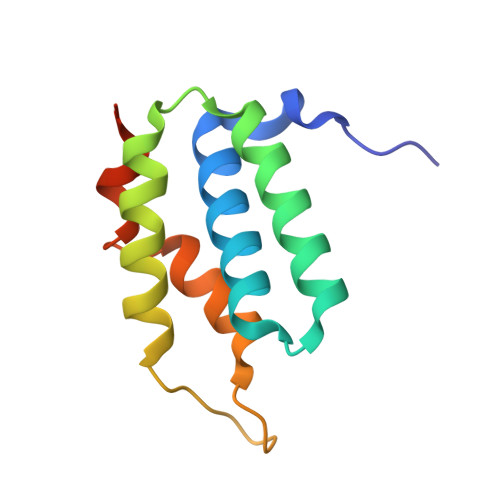
Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein.
Zhang, H., Zhang, H., Gao, Z.Q., Wang, W.J., Liu, G.F., Xu, J.H., Su, X.D., Dong, Y.H.(2013) J Biological Chem 288: 5928-5939
- PubMed: 23288853
- DOI: https://doi.org/10.1074/jbc.M112.434357
- Primary Citation of Related Structures:
4HFF, 4HFK, 4HFL - PubMed Abstract:
The type VI secretion system (T6SS), a multisubunit needle-like apparatus, has recently been found to play a role in interspecies interactions. The gram-negative bacteria harboring T6SS (donor) deliver the effectors into their neighboring cells (recipient) to kill them. Meanwhile, the cognate immunity proteins were employed to protect the donor cells against the toxic effectors. Tae4 (type VI amidase effector 4) and Tai4 (type VI amidase immunity 4) are newly identified T6SS effector-immunity pairs. Here, we report the crystal structures of Tae4 from Enterobacter cloacae and Tae4-Tai4 complexes from both E. cloacae and Salmonella typhimurium. Tae4 acts as a DL-endopeptidase and displays a typical N1pC/P60 domain. Unlike Tsi1 (type VI secretion immunity 1), Tai4 is an all-helical protein and forms a dimer in solution. The small angle x-ray scattering study combined with the analytical ultracentrifugation reveal that the Tae4-Tai4 complex is a compact heterotetramer that consists of a Tai4 dimer and two Tae4 molecules in solution. Structure-based mutational analysis of the Tae4-Tai4 interface shows that a helix (α3) of one subunit in dimeric Tai4 plays a major role in binding of Tae4, whereas a protruding loop (L4) in the other subunit is mainly responsible for inhibiting Tae4 activity. The inhibition process requires collaboration between the Tai4 dimer. These results reveal a novel and unique inhibition mechanism in effector-immunity pairs and suggest a new strategy to develop antipathogen drugs.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, No. 5 Yiheyuan Road, Beijing 100871, China.
Organizational Affiliation: