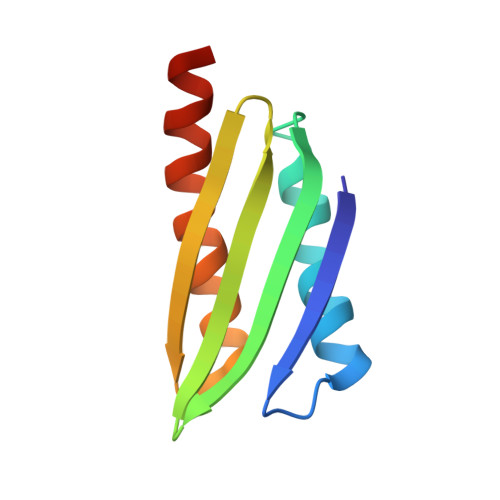
Structure of Vibrio cholerae ribosome hibernation promoting factor.
De Bari, H., Berry, E.A.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 228-236
- PubMed: 23519794
- DOI: https://doi.org/10.1107/S1744309113000961
- Primary Citation of Related Structures:
4HEI - PubMed Abstract:
The X-ray crystal structure of ribosome hibernation promoting factor (HPF) from Vibrio cholerae is presented at 2.0 Å resolution. The crystal was phased by two-wavelength MAD using cocrystallized cobalt. The asymmetric unit contained two molecules of HPF linked by four Co atoms. The metal-binding sites observed in the crystal are probably not related to biological function. The structure of HPF has a typical β-α-β-β-β-α fold consistent with previous structures of YfiA and HPF from Escherichia coli. Comparison of the new structure with that of HPF from E. coli bound to the Thermus thermophilus ribosome [Polikanov et al. (2012), Science, 336, 915-918] shows that no significant structural changes are induced in HPF by binding.
- Biochemistry and Molecular Biology, SUNY Upstate Medical University, 750 E. Adams Avenue, Syracuse, NY 13210, USA.
Organizational Affiliation: