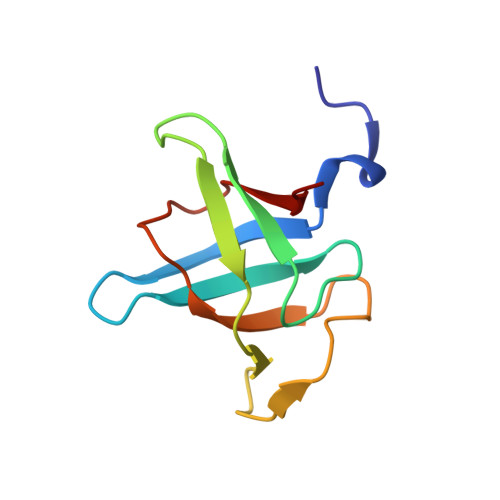
Ultratight crystal packing of a 10 kDa protein.
Trillo-Muyo, S., Jasilionis, A., Domagalski, M.J., Chruszcz, M., Minor, W., Kuisiene, N., Arolas, J.L., Sola, M., Gomis-Ruth, F.X.(2013) Acta Crystallogr D Biol Crystallogr 69: 464-470
- PubMed: 23519421
- DOI: https://doi.org/10.1107/S0907444912050135
- Primary Citation of Related Structures:
4HE5, 4HE6 - PubMed Abstract:
While small organic molecules generally crystallize forming tightly packed lattices with little solvent content, proteins form air-sensitive high-solvent-content crystals. Here, the crystallization and full structure analysis of a novel recombinant 10 kDa protein corresponding to the C-terminal domain of a putative U32 peptidase are reported. The orthorhombic crystal contained only 24.5% solvent and is therefore among the most tightly packed protein lattices ever reported.
- Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona, Spanish Research Council CSIC, Barcelona Science Park, c/Baldiri Reixac 15-21, 08028 Barcelona, Spain.
Organizational Affiliation: