A Different Fold with an Integrin-Binding Loop Specialized for Flexibility in Mucosal Addressin Cell Adhesion Molecule-1
Springer, T., Yu, Y., Zhu, J., Wang, J.-H., Huang, P.-S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PF-547659 heavy chain | A [auth H], D [auth M] | 229 | Homo sapiens | Mutation(s): 0 | 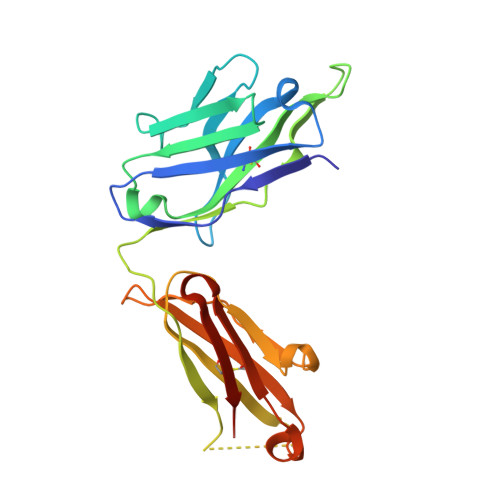 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PF-547659 light chain | B [auth L], E [auth N] | 219 | Homo sapiens | Mutation(s): 0 | 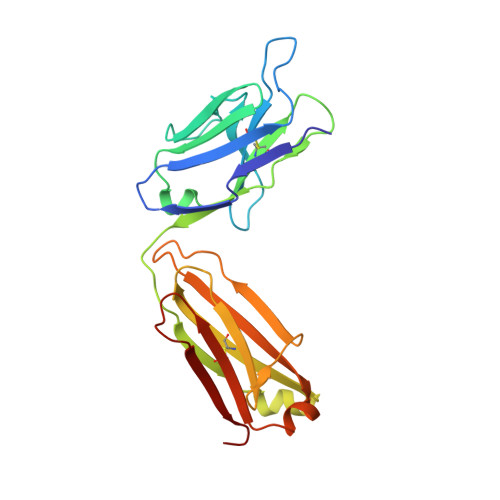 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mucosal addressin cell adhesion molecule 1 | C [auth A], F [auth B] | 209 | Homo sapiens | Mutation(s): 1 Gene Names: MADCAM1 | 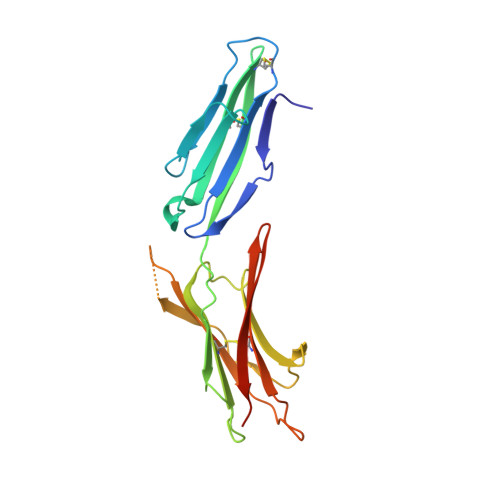 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13477 (Homo sapiens) Explore Q13477 Go to UniProtKB: Q13477 | |||||
PHAROS: Q13477 GTEx: ENSG00000099866 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13477 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.94 | α = 90 |
| b = 112.26 | β = 89.95 |
| c = 157.92 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASER | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| SCALA | data scaling |