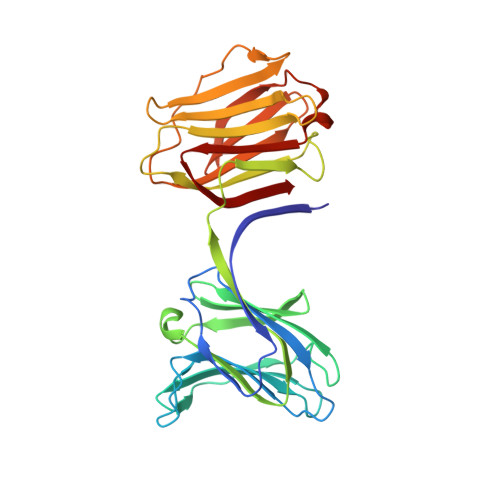
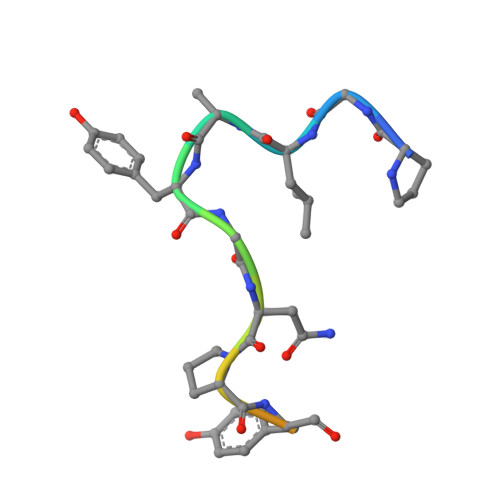
Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Kim, B.W., Hong, S.B., Kim, J.H., Kwon, D.H., Song, H.K.(2013) Nat Commun 4: 1613-1613
- PubMed: 23511477
- DOI: https://doi.org/10.1038/ncomms2606
- Primary Citation of Related Structures:
4HAN - PubMed Abstract:
Infectious bacteria are cleared from mammalian cells by host autophagy in combination with other upstream cellular components, such as the autophagic receptor NDP52 and sugar receptor galectin-8. However, the detailed molecular basis of the interaction between these two receptors remains to be elucidated. Here, we report the biochemical characterization of both NDP52 and galectin-8 as well as the crystal structure of galectin-8 complexed with an NDP52 peptide. The unexpected observation of nicotinamide adenine dinucleotide located at the carbohydrate-binding site expands our knowledge of the sugar-binding specificity of galectin-8. The NDP52-galectin-8 complex structure explains the key determinants for recognition on both receptors and defines a special orientation of N- and C-terminal carbohydrate recognition domains of galectin-8. Dimeric NDP52 forms a ternary complex with two monomeric galectin-8 molecules as well as two LC3C molecules. These results lay the groundwork for understanding how host cells target bacterial pathogens for autophagy.
- Division of Life Sciences, School of Life Sciences and Biotechnology, Korea University, Anam-Dong, Seongbuk-Gu, Seoul, Korea.
Organizational Affiliation: