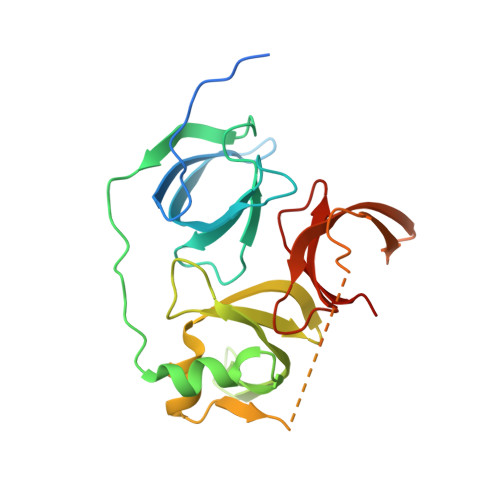
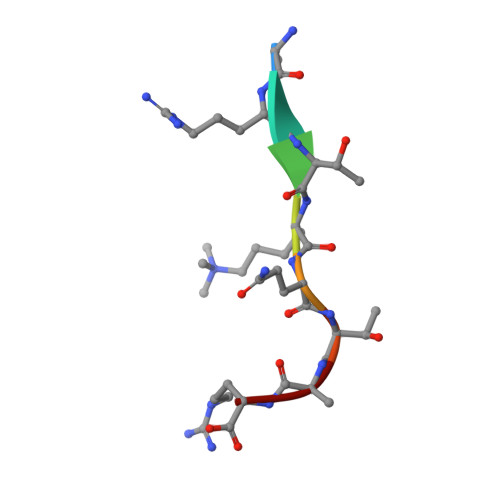
Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Yang, N., Wang, W., Wang, Y., Wang, M., Zhao, Q., Rao, Z., Zhu, B., Xu, R.M.(2012) Proc Natl Acad Sci U S A 109: 17954-17959
- PubMed: 23077255
- DOI: https://doi.org/10.1073/pnas.1208517109
- Primary Citation of Related Structures:
4H75 - PubMed Abstract:
Recognition of methylated histone tail lysine residues by tudor domains plays important roles in epigenetic control of gene expression and DNA damage response. Previous studies revealed the binding of methyllysine in a cage of aromatic residues, but the molecular mechanism by which the sequence specificity for surrounding histone tail residues is achieved remains poorly understood. In the crystal structure of a trimethylated histone H3 lysine 4 (H3K4) peptide bound to the tudor-like domains of Spindlin1 presented here, an atypical mode of methyllysine recognition by an aromatic pocket of Spindlin1 is observed. Furthermore, the histone sequence is recognized in a distinct manner involving the amino terminus and a pair of arginine residues of histone H3, and disruption of the binding impaired stimulation of pre-RNA expression by Spindlin1. Our analysis demonstrates considerable diversities of methyllysine recognition and sequence-specific binding of histone tails by tudor domains, and the revelation furthers the understanding of tudor domain proteins in deciphering epigenetic marks on histone tails.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: