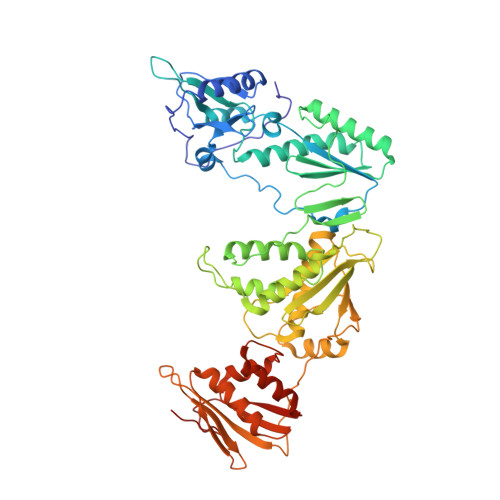
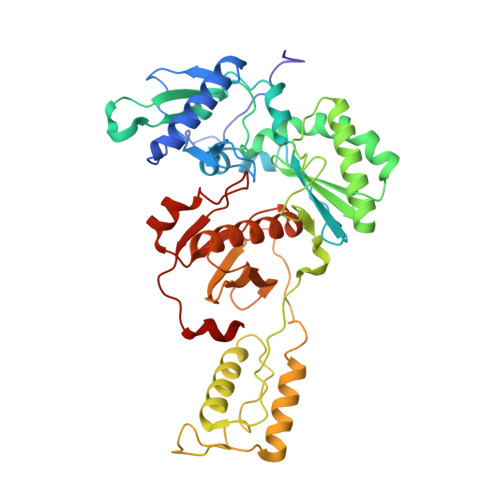
Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
Frey, K.M., Bollini, M., Mislak, A.C., Cisneros, J.A., Gallardo-Macias, R., Jorgensen, W.L., Anderson, K.S.(2012) J Am Chem Soc 134: 19501-19503
- PubMed: 23163887
- DOI: https://doi.org/10.1021/ja3092642
- Primary Citation of Related Structures:
4H4M, 4H4O - PubMed Abstract:
X-ray crystal structures at 2.9 Å resolution are reported for two complexes of catechol diethers with HIV-1 reverse transcriptase. The results help elucidate the structural origins of the extreme antiviral activity of the compounds. The possibility of halogen bonding between the inhibitors and Pro95 is addressed. Structural analysis reveals key interactions with conserved residues P95 and W229 of importance for design of inhibitors with high potency and favorable resistance profiles.
- Department of Chemistry, Yale University, New Haven, Connecticut 06520-8107, United States.
Organizational Affiliation: