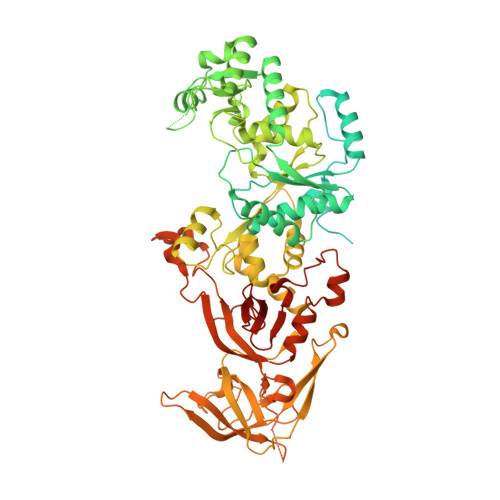
The Structure of Yeast Glutaminyl-tRNA Synthetase and Modeling of Its Interaction with tRNA.
Grant, T.D., Luft, J.R., Wolfley, J.R., Snell, M.E., Tsuruta, H., Corretore, S., Quartley, E., Phizicky, E.M., Grayhack, E.J., Snell, E.H.(2013) J Mol Biology 425: 2480-2493
- PubMed: 23583912
- DOI: https://doi.org/10.1016/j.jmb.2013.03.043
- Primary Citation of Related Structures:
4H3S - PubMed Abstract:
Eukaryotic glutaminyl-tRNA synthetase (GlnRS) contains an appended N-terminal domain (NTD) whose precise function is unknown. Although GlnRS structures from two prokaryotic species are known, no eukaryotic GlnRS structure has been reported. Here we present the first crystallographic structure of yeast GlnRS, finding that the structure of the C-terminal domain is highly similar to Escherichia coli GlnRS but that 214 residues, including the NTD, are crystallographically disordered. We present a model of the full-length enzyme in solution, using the structures of the C-terminal domain, and the isolated NTD, with small-angle X-ray scattering data of the full-length molecule. We proceed to model the enzyme bound to tRNA, using the crystallographic structures of GatCAB and GlnRS-tRNA complex from bacteria. We contrast the tRNA-bound model with the tRNA-free solution state and perform molecular dynamics on the full-length GlnRS-tRNA complex, which suggests that tRNA binding involves the motion of a conserved hinge in the NTD.
- Hauptman Woodward Medical Research Institute, 700 Ellicott Street, Buffalo, NY 14203, USA.
Organizational Affiliation: