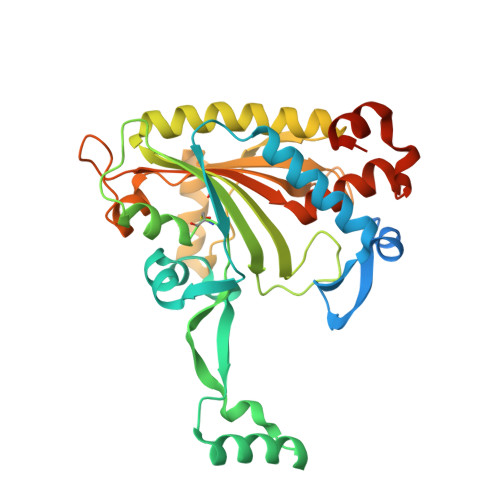
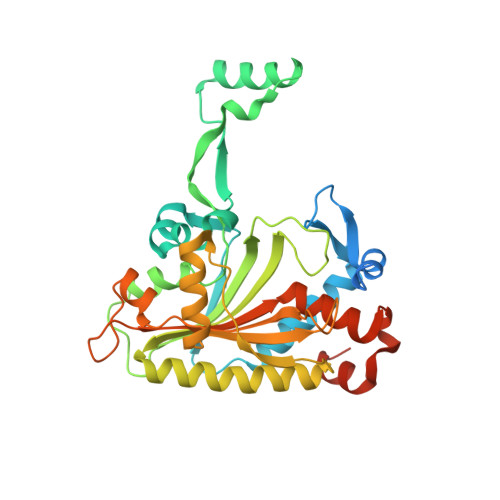
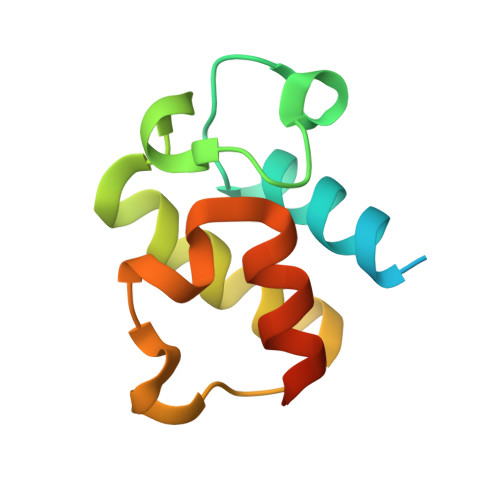
Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Mocibob, M., Ivic, N., Luic, M., Weygand-Durasevic, I.(2013) Structure 21: 614-626
- PubMed: 23541895
- DOI: https://doi.org/10.1016/j.str.2013.02.017
- Primary Citation of Related Structures:
4H2S, 4H2T, 4H2U, 4H2V, 4H2W, 4H2X, 4H2Y - PubMed Abstract:
Amino acid:[carrier protein] ligases (aa:CP ligases) are recently discovered enzymes that are highly similar to class II aminoacyl-tRNA synthetases (aaRSs). However, while aaRSs aminoacylate tRNA and supply building blocks for ribosomal translation, aa:CP ligases transfer activated amino acids to the phosphopantetheine group of small carrier proteins. We have solved the crystal structure of an aa:CP ligase complexed with the carrier protein (CP). The CP prosthetic group enters the active site from a different direction than tRNA in class II aaRS complexes through an idiosyncratic tunnel. CP binds to aa:CP ligase in a fundamentally different manner compared to tRNA binding by structurally closely related aaRSs. Based on crystallographic analysis, an enzyme of altered CP specificity was designed, and the mechanism of amino acid transfer to the prosthetic group was proposed. The presented study reveals how a conserved class II aaRS catalytic core can adapt to another function through minor structural alterations.
- Department of Chemistry, Faculty of Science, University of Zagreb, Horvatovac 102a, 10 000 Zagreb, Croatia.
Organizational Affiliation: