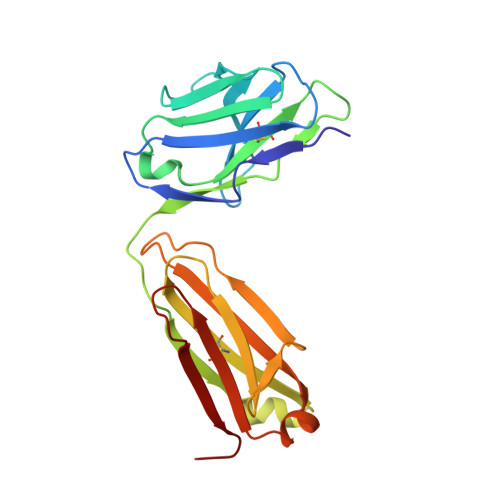
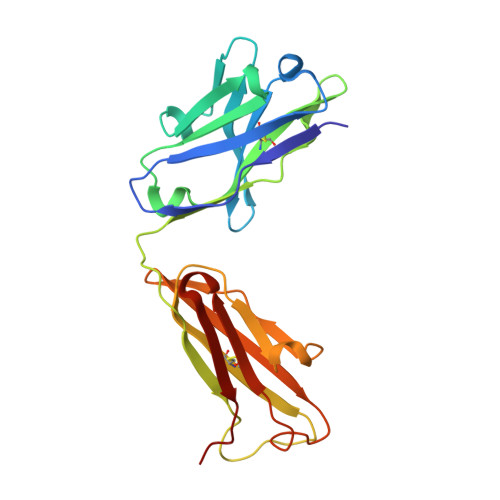
Crystal structure and computational modeling of the fab fragment from a protective anti-ricin monoclonal antibody.
Zhao, Z., Worthylake, D., Lecour, L., Maresh, G.A., Pincus, S.H.(2012) PLoS One 7: e52613-e52613
- PubMed: 23285112
- DOI: https://doi.org/10.1371/journal.pone.0052613
- Primary Citation of Related Structures:
4H20 - PubMed Abstract:
Many antibody crystal structures have been solved. Structural modeling programs have been developed that utilize this information to predict 3-D structures of an antibody based upon its sequence. Because of the problem of self-reference, the accuracy and utility of these predictions can only be tested when a new structure has not yet been deposited in the Protein Data Bank. We have solved the crystal structure of the Fab fragment of RAC18, a protective anti-ricin mAb, to 1.9 Å resolution. We have also modeled the Fv structure of RAC18 using publicly available Ab modeling tools Prediction of Immunoglobulin Structures (PIGS), RosettaAntibody, and Web Antibody Modeling (WAM). The model structures underwent energy minimization. We compared results to the crystal structure on the basis of root-mean-square deviation (RMSD), template modeling score (TM-score), Z-score, and MolProbity analysis. The crystal structure showed a pocket formed mainly by AA residues in each of the heavy chain complementarity determining regions (CDRs). There were differences between the crystal structure and structures predicted by the modeling tools, particularly in the CDRs. There were also differences among the predicted models, although the differences were small and within experimental error. No one modeling program was clearly superior to the others. In some cases, choosing structures based only on sequence homology to the crystallized Ab yielded RMSDs comparable to the models. Molecular modeling programs accurately predict the structure of most regions of antibody variable domains of RAC18. The hypervariable CDRs proved most difficult to model, particularly H chain CDR3. Because CDR3 is most often involved in contact with antigen, this defect must be considered when using models to identify potential contacts between antibody and antigen. Because this study represents only a single case, the results cannot be generalized. Rather they highlight the utility and limitations of modeling programs.
- The Research Institute for Children, Children's Hospital, New Orleans, Louisiana, United States of America.
Organizational Affiliation: