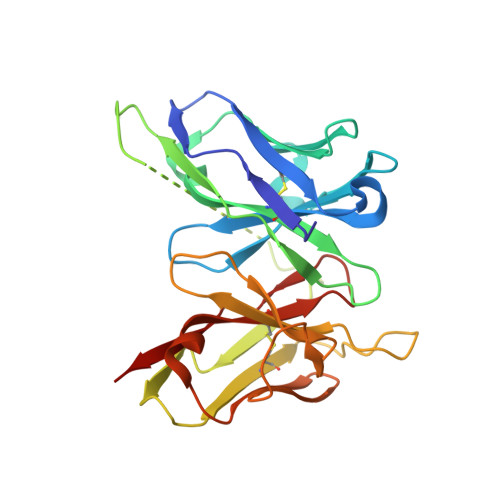
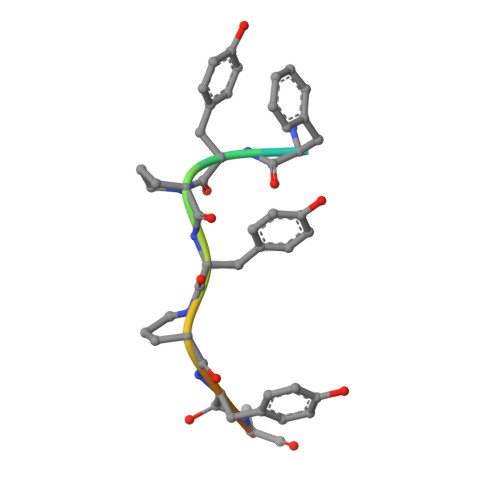
Structural evaluation of a mimicry-recognizing paratope: plasticity in antigen-antibody interactions manifests in molecular mimicry.
Tapryal, S., Gaur, V., Kaur, K.J., Salunke, D.M.(2013) J Immunol 191: 456-463
- PubMed: 23733869
- DOI: https://doi.org/10.4049/jimmunol.1203260
- Primary Citation of Related Structures:
4H0G, 4H0H, 4H0I - PubMed Abstract:
Molecular mimicry manifests antagonistically with respect to the specificity of immune recognition. However, it often occurs because different Ags share surface topologies in terms of shape or chemical nature. It also occurs when a flexible paratope accommodates dissimilar Ags by adjusting structural features according to the antigenic epitopes or differential positioning in the Ag combining site. Toward deciphering the structural basis of molecular mimicry, mAb 2D10 was isolated from a maturing immune response elicited against methyl α-d-mannopyranoside and also bound equivalently to a dodecapeptide. The physicochemical evidence of this carbohydrate-peptide mimicry in the case of mAb 2D10 had been established earlier. These studies had strongly suggested direct involvement of a flexible paratope in the observed mimicry. Surprisingly, comparison of the Ag-free structure of single-chain variable fragment 2D10 with those bound to sugar and peptide Ags revealed a conformationally invariant state of the Ab while binding to chemically and structurally disparate Ags. This equivalent binding of the two dissimilar Ags was through mutually independent interactions, demonstrating functional equivalence in the absence of structural correlation. Thus, existence of a multispecific, mature Ab in the secondary immune response was evident, as was the plasticity in the interactions while accommodating topologically diverse Ags. Although our data highlight the structural basis of receptor multispecificity, they also illustrate mechanisms adopted by the immune system to neutralize the escape mutants generated during pathogenic insult.
- National Institute of Immunology, New Delhi 110067, India.
Organizational Affiliation: