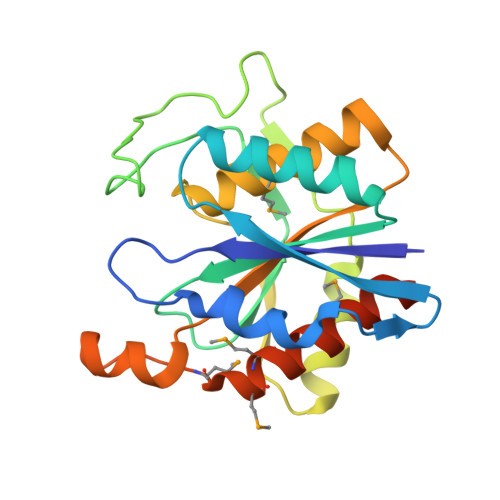
Crystal Structure of a Pyrrolidone-carboxylate peptidase 1 (target ID NYSGRC-012831) from Xenorhabdus bovienii SS-2004 (CASP Target)
Ghosh, A., Bhoshle, R., Toro, R., Gizzi, A., Hillerich, B., Seidel, R., Almo, S.C., New York Structural Genomics Research ConsortiumTo be published.