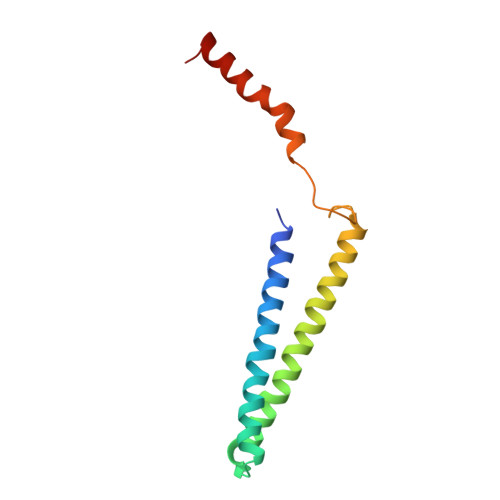
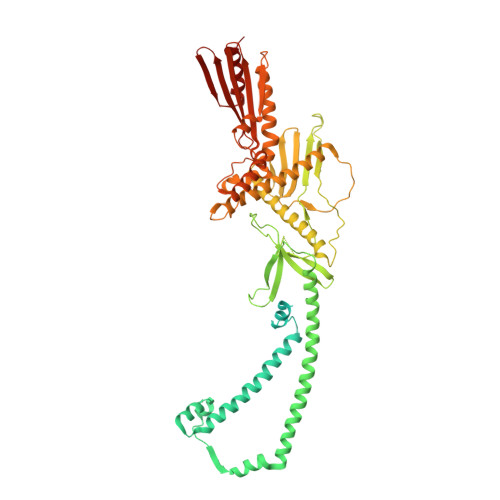
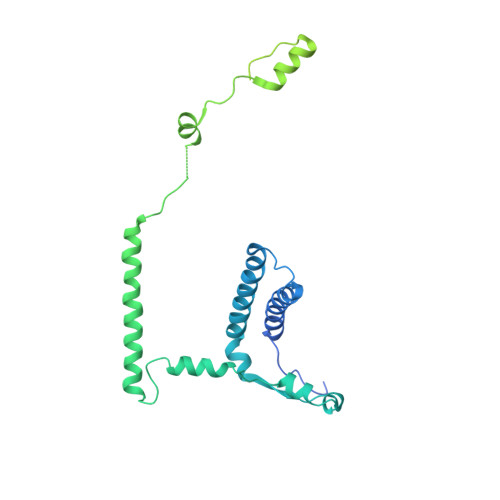
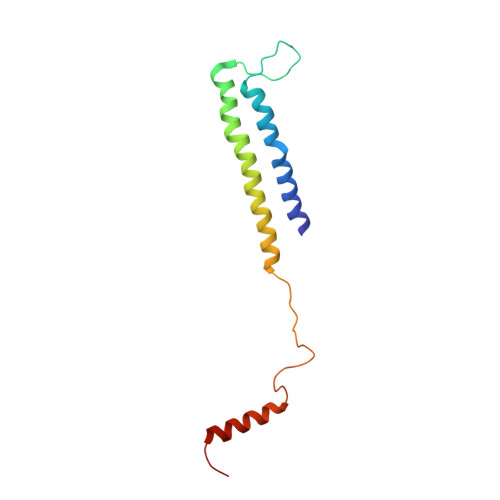
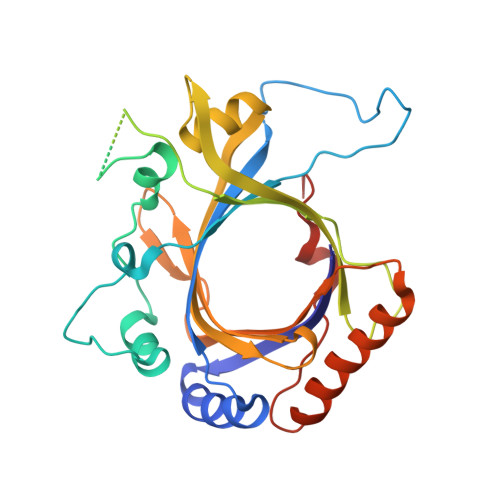
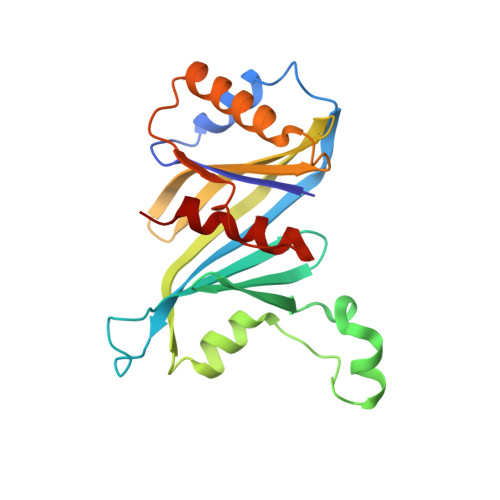
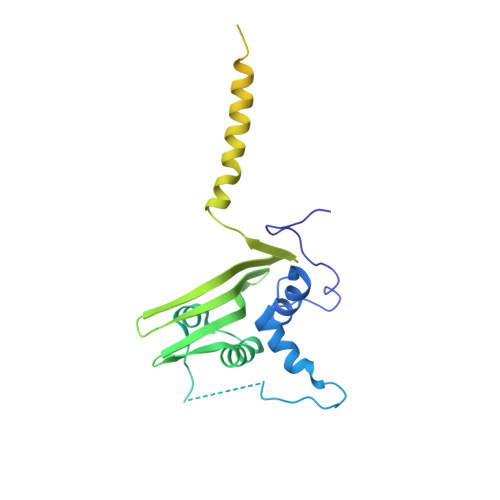
Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II.
Robinson, P.J., Bushnell, D.A., Trnka, M.J., Burlingame, A.L., Kornberg, R.D.(2012) Proc Natl Acad Sci U S A 109: 17931-17935
- PubMed: 23071300
- DOI: https://doi.org/10.1073/pnas.1215241109
- Primary Citation of Related Structures:
4GWP, 4GWQ - PubMed Abstract:
The X-ray crystal structure of the Head module, one-third of the Mediator of transcriptional regulation, has been determined as a complex with the C-terminal domain (CTD) of RNA polymerase II. The structure reveals multiple points of interaction with an extended conformation of the CTD; it suggests a basis for regulation by phosphorylation of the CTD. Biochemical studies show a requirement for Mediator-CTD interaction for transcription.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: