Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15.
Ring, A.M., Lin, J.X., Feng, D., Mitra, S., Rickert, M., Bowman, G.R., Pande, V.S., Li, P., Moraga, I., Spolski, R., Ozkan, E., Leonard, W.J., Garcia, K.C.(2012) Nat Immunol 13: 1187-1195
- PubMed: 23104097
- DOI: https://doi.org/10.1038/ni.2449
- Primary Citation of Related Structures:
4GS7 - PubMed Abstract:
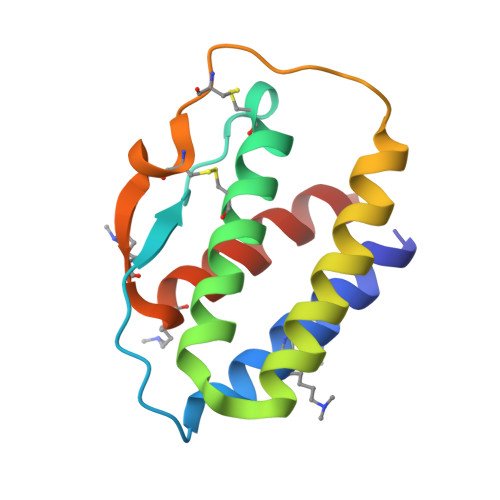
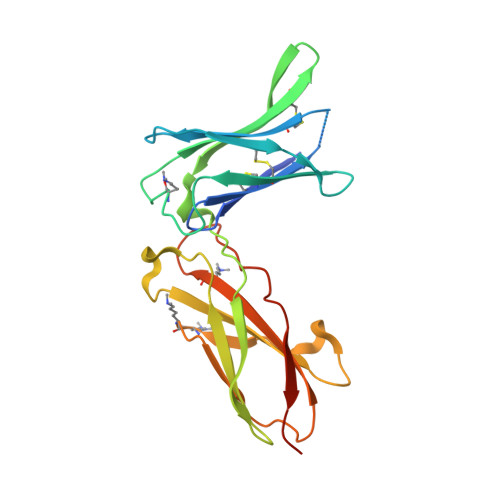
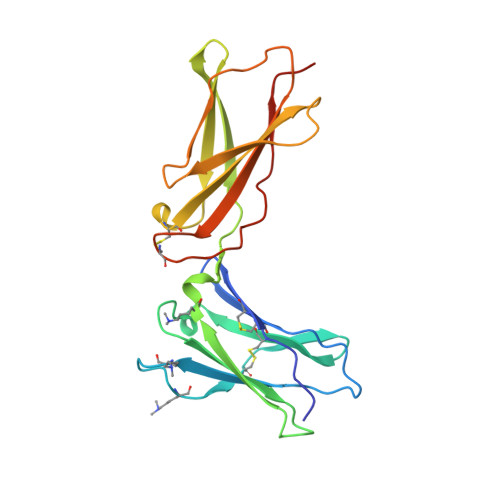
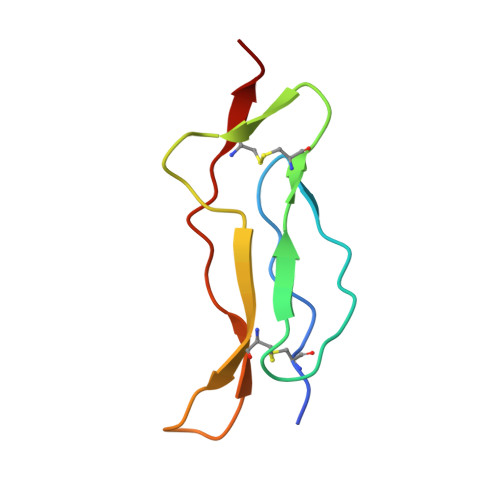
Interleukin 15 (IL-15) and IL-2 have distinct immunological functions even though both signal through the receptor subunit IL-2Rβ and the common γ-chain (γ(c)). Here we found that in the structure of the IL-15-IL-15Rα-IL-2Rβ-γ(c) quaternary complex, IL-15 binds to IL-2Rβ and γ(c) in a heterodimer nearly indistinguishable from that of the IL-2-IL-2Rα-IL-2Rβ-γ(c) complex, despite their different receptor-binding chemistries. IL-15Rα substantially increased the affinity of IL-15 for IL-2Rβ, and this allostery was required for IL-15 trans signaling. Consistent with their identical IL-2Rβ-γ(c) dimer geometries, IL-2 and IL-15 showed similar signaling properties in lymphocytes, with any differences resulting from disparate receptor affinities. Thus, IL-15 and IL-2 induced similar signals, and the cytokine specificity of IL-2Rα versus IL-15Rα determined cellular responsiveness. Our results provide new insights for the development of specific immunotherapeutics based on IL-15 or IL-2.
- Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California, USA.
Organizational Affiliation: