Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells.
Etchin, J., Sun, Q., Kentsis, A., Farmer, A., Zhang, Z.C., Sanda, T., Mansour, M.R., Barcelo, C., McCauley, D., Kauffman, M., Shacham, S., Christie, A.L., Kung, A.L., Rodig, S.J., Chook, Y.M., Look, A.T.(2013) Leukemia 27: 66-74
- PubMed: 22847027
- DOI: https://doi.org/10.1038/leu.2012.219
- Primary Citation of Related Structures:
4GPT - PubMed Abstract:
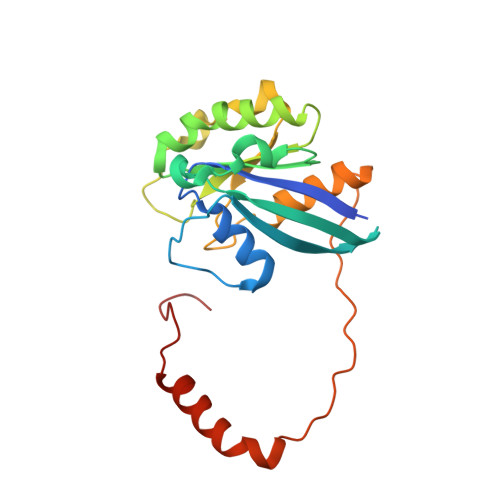
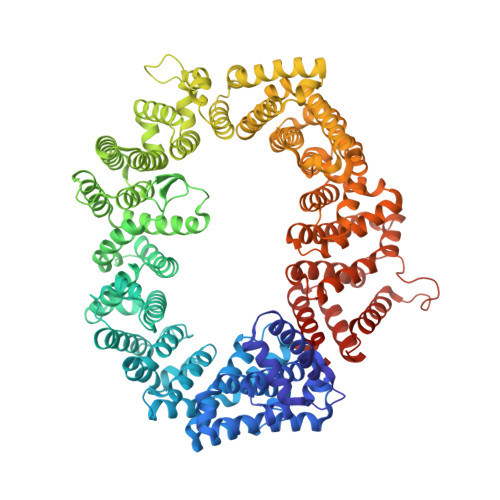
Drugs that target the chief mediator of nuclear export, chromosome region maintenance 1 protein (CRM1) have potential as therapeutics for leukemia, but existing CRM1 inhibitors show variable potencies and a broad range of cytotoxic effects. Here, we report the structural analysis and antileukemic activity of a new generation of small-molecule inhibitors of CRM1. Designated selective inhibitors of nuclear export (SINE), these compounds were developed using molecular modeling to screen a small virtual library of compounds against the nuclear export signal (NES) groove of CRM1. The 2.2-Å crystal structure of the CRM1-Ran-RanBP1 complex bound to KPT-251, a representative molecule of this class of inhibitors, shows that the drug occupies part of the groove in CRM1 that is usually occupied by the NES, but penetrates much deeper into the groove and blocks CRM1-directed protein export. SINE inhibitors exhibit potent antileukemic activity, inducing apoptosis at nanomolar concentrations in a panel of 14 human acute myeloid leukemia (AML) cell lines representing different molecular subtypes of the disease. When administered orally to immunodeficient mice engrafted with human AML cells, KPT-251 had potent antileukemic activity with negligible toxicity to normal hematopoietic cells. Thus, KPT-SINE CRM1 antagonists represent a novel class of drugs that warrant further testing in AML patients.
- Department of Pediatric Oncology, Dana-Farber Cancer Institute, Division of Hematology/Oncology, Children's Hospital Boston, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: