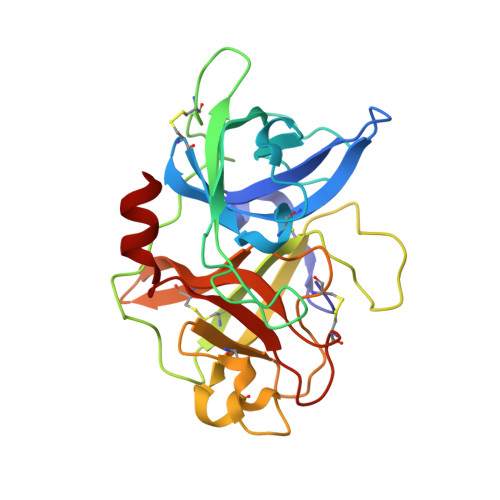
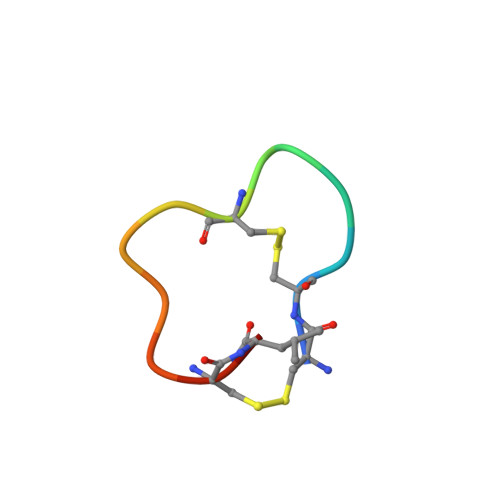
Bicyclic Peptide Ligands Pulled out of Cysteine-Rich Peptide Libraries.
Chen, S., Rentero Rebollo, I., Buth, S.A., Morales-Sanfrutos, J., Touati, J., Leiman, P.G., Heinis, C.(2013) J Am Chem Soc 135: 6562-6569
- PubMed: 23560397
- DOI: https://doi.org/10.1021/ja400461h
- Primary Citation of Related Structures:
4GLY - PubMed Abstract:
Bicyclic peptide ligands were found to have good binding affinity and target specificity. However, the method applied to generate bicyclic ligands based on phage-peptide alkylation is technically complex and limits its application to specialized laboratories. Herein, we report a method that involves a simpler and more robust procedure that additionally allows screening of structurally more diverse bicyclic peptide libraries. In brief, phage-encoded combinatorial peptide libraries of the format X(m)CX(n)CX(o)CX(p) are oxidized to connect two pairs of cysteines (C). This allows the generation of 3 × (m + n + o + p) different peptide topologies because the fourth cysteine can appear in any of the (m + n + o + p) randomized amino acid positions (X). Panning of such libraries enriched strongly peptides with four cysteines and yielded tight binders to protein targets. X-ray structure analysis revealed an important structural role of the disulfide bridges. In summary, the presented approach offers facile access to bicyclic peptide ligands with good binding affinities.
- Institute of Chemical Sciences and Engineering, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland.
Organizational Affiliation: