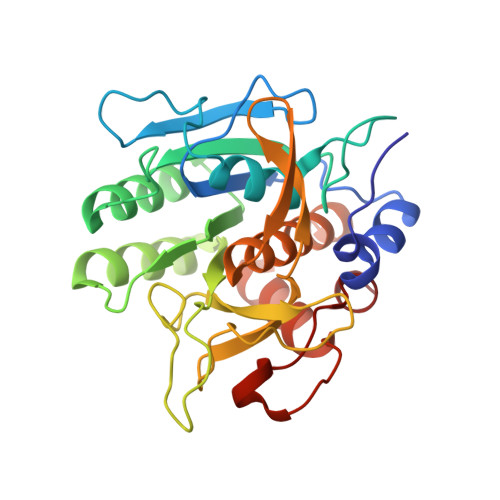
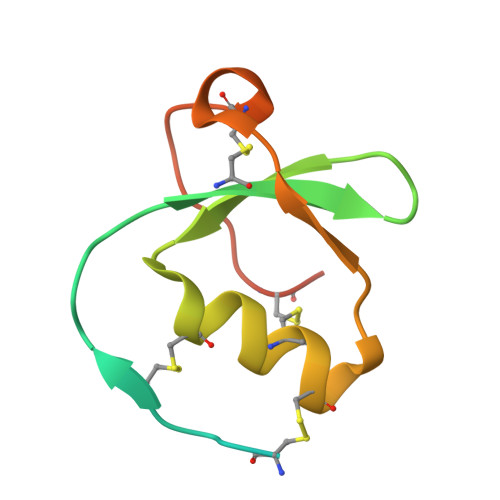
Crystal structure of greglin, a novel non-classical Kazal inhibitor, in complex with subtilisin
Derache, C., Epinette, C., Roussel, A., Gabant, G., Cadene, M., Korkmaz, B., Gauthier, F., Kellenberger, C.(2012) FEBS J 279: 4466-4478
- PubMed: 23075397
- DOI: https://doi.org/10.1111/febs.12033
- Primary Citation of Related Structures:
4GI3 - PubMed Abstract:
Greglin is an 83-residue serine protease inhibitor purified from the ovaries of the locust Schistocerca gregaria. Greglin is a strong inhibitor of subtilisin and human neutrophil elastase, acting at sub-nanomolar and nanomolar concentrations, respectively; it also inhibits neutrophil cathepsin G, α-chymotrypsin and porcine pancreatic elastase, but to a lesser extent. In the present study, we show that greglin resists denaturation at high temperature (95 °C) and after exposure to acetonitrile and acidic or basic pH. Greglin is composed of two domains consisting of residues 1-20 and 21-83. Mass spectrometry indicates that the N-terminal domain (1-20) is post-translationally modified by phosphorylations at three sites and probably contains a glycosylation site. The crystal structure of the region of greglin comprising residues 21-78 in complex with subtilisin was determined at 1.75 Å resolution. Greglin represents a novel member of the non-classical Kazal inhibitors, as it has a unique additional C-terminal region (70-83) connected to the core of the molecule via a supplementary disulfide bond. The stability of greglin was compared with that of an ovomucoid inhibitor. The thermostability and inhibitory specificity of greglin are discussed in light of its structure. In particular, we propose that the C-terminal region is responsible for non-favourable interactions with the autolysis loop (140-loop) of serine proteases of the chymotrypsin family, and thus governs specificity. The atomic coordinates and structure factors for the greglin-subtilisin complex have been deposited with the RCSB Protein Data Bank under accession number 4GI3. Greglin and Subtilisin Carlsberg bind by X-ray crystallography (View interaction).
- Centre de Biophysique Moléculaire, UPR 4301 CNRS conventionnée avec l'Université d'Orléans, Orléans Cedex 2, France.
Organizational Affiliation: