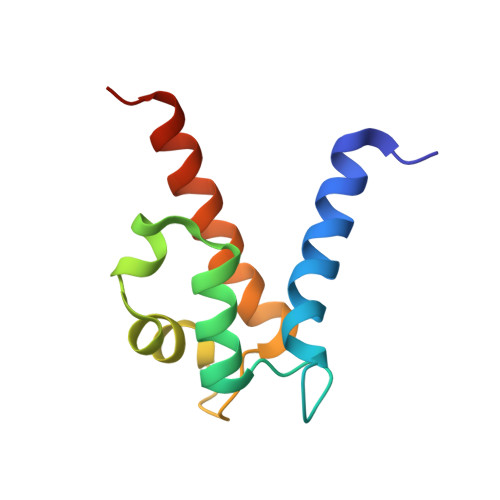
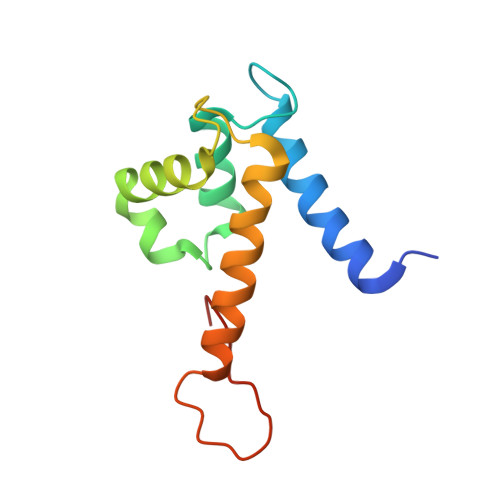
Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens.
Damo, S.M., Kehl-Fie, T.E., Sugitani, N., Holt, M.E., Rathi, S., Murphy, W.J., Zhang, Y., Betz, C., Hench, L., Fritz, G., Skaar, E.P., Chazin, W.J.(2013) Proc Natl Acad Sci U S A 110: 3841-3846
- PubMed: 23431180
- DOI: https://doi.org/10.1073/pnas.1220341110
- Primary Citation of Related Structures:
4GGF - PubMed Abstract:
The S100A8/S100A9 heterodimer calprotectin (CP) functions in the host response to pathogens through a mechanism termed "nutritional immunity." CP binds Mn(2+) and Zn(2+) with high affinity and starves bacteria of these essential nutrients. Combining biophysical, structural, and microbiological analysis, we identified the molecular basis of Mn(2+) sequestration. The asymmetry of the CP heterodimer creates a single Mn(2+)-binding site from six histidine residues, which distinguishes CP from all other Mn(2+)-binding proteins. Analysis of CP mutants with altered metal-binding properties revealed that, despite both Mn(2+) and Zn(2+) being essential metals, maximal growth inhibition of multiple bacterial pathogens requires Mn(2+) sequestration. These data establish the importance of Mn(2+) sequestration in defense against infection, explain the broad-spectrum antimicrobial activity of CP relative to other S100 proteins, and clarify the impact of metal depletion on the innate immune response to infection.
- Department of Biochemistry, and Center for Structural Biology, Vanderbilt University, Nashville, TN 37232-8725, USA.
Organizational Affiliation: