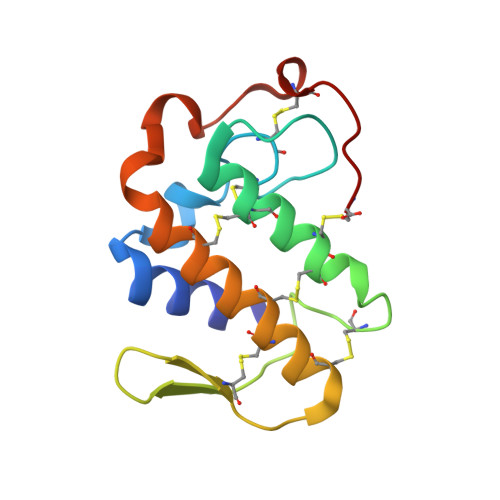
Design of peptide inhibitors of phospholipase A2: crystal Structure of phospholipase A2 complexed with a designed tetrapeptide Val - Ilu- Ala - Lys at 2.7 A resolution
Shukla, P.K., Sinha, M., Dey, S., Kaur, P., Sharma, S., Singh, T.P.To be published.