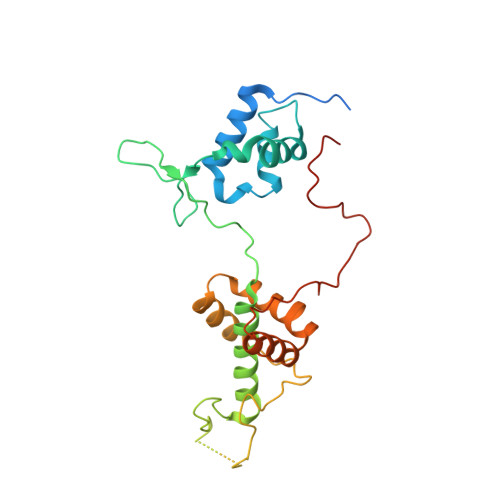
Effect of Rap1 binding on DNA distortion and potassium permanganate hypersensitivity.
Le Bihan, Y.V., Matot, B., Pietrement, O., Giraud-Panis, M.J., Gasparini, S., Le Cam, E., Gilson, E., Sclavi, B., Miron, S., Le Du, M.H.(2013) Acta Crystallogr D Biol Crystallogr 69: 409-419
- PubMed: 23519416
- DOI: https://doi.org/10.1107/S0907444912049311
- Primary Citation of Related Structures:
4GFB - PubMed Abstract:
Repressor activator protein 1 (Rap1) is an essential factor involved in transcription and telomere stability in the budding yeast Saccharomyces cerevisiae. Its interaction with DNA causes hypersensitivity to potassium permanganate, suggesting local DNA melting and/or distortion. In this study, various Rap1-DNA crystal forms were obtained using specifically designed crystal screens. Analysis of the DNA conformation showed that its distortion was not sufficient to explain the permanganate reactivity. However, anomalous data collected at the Mn edge using a Rap1-DNA crystal soaked in potassium permanganate solution indicated that the DNA conformation in the crystal was compatible with interaction with permanganate ions. Sequence-conservation analysis revealed that double-Myb-containing Rap1 proteins all carry a fully conserved Arg580 at a position that may favour interaction with permanganate ions, although it is not involved in the hypersensitive cytosine distortion. Permanganate reactivity assays with wild-type Rap1 and the Rap1[R580A] mutant demonstrated that Arg580 is essential for hypersensitivity. AFM experiments showed that wild-type Rap1 and the Rap1[R580A] mutant interact with DNA over 16 successive binding sites, leading to local DNA stiffening but not to accumulation of the observed local distortion. Therefore, Rap1 may cause permanganate hypersensitivity of DNA by forming a pocket between the reactive cytosine and Arg580, driving the permanganate ion towards the C5-C6 bond of the cytosine.
- CEA/DSV/IBiTec-S/SB2SM, Laboratoire de Biologie Structurale et Radiobiologie, CNRS UMR 8221, University Paris-Sud, Bâtiment 144, CE Saclay, 91191 Gif-sur-Yvette, France.
Organizational Affiliation: