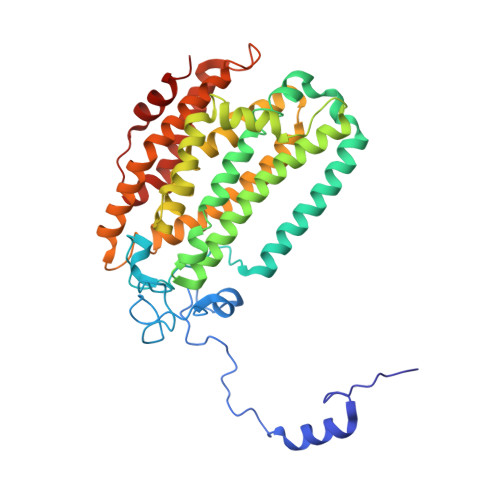
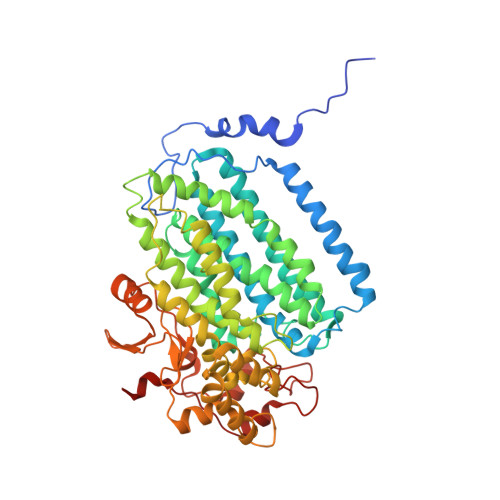
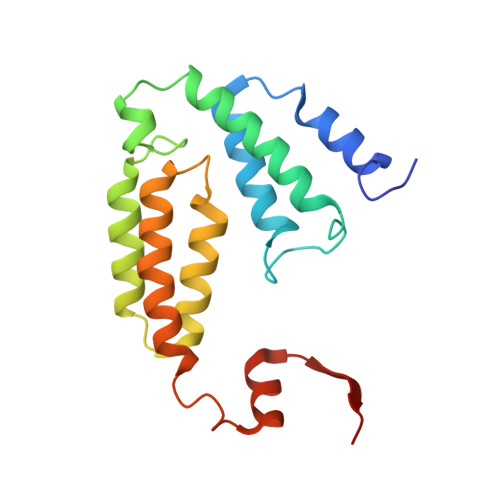
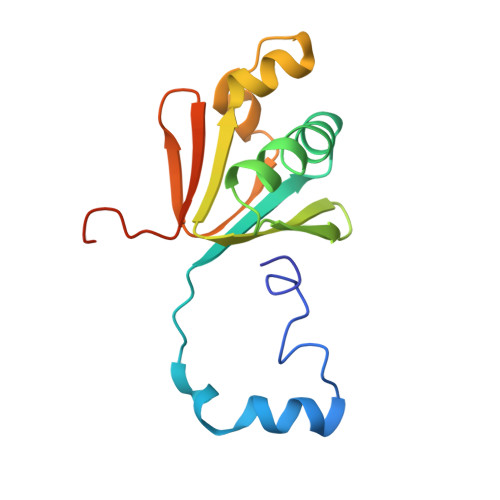
Control of substrate access to the active site in methane monooxygenase.
Lee, S.J., McCormick, M.S., Lippard, S.J., Cho, U.S.(2013) Nature 494: 380-384
- PubMed: 23395959
- DOI: https://doi.org/10.1038/nature11880
- Primary Citation of Related Structures:
4GAM - PubMed Abstract:
Methanotrophs consume methane as their major carbon source and have an essential role in the global carbon cycle by limiting escape of this greenhouse gas to the atmosphere. These bacteria oxidize methane to methanol by soluble and particulate methane monooxygenases (MMOs). Soluble MMO contains three protein components, a 251-kilodalton hydroxylase (MMOH), a 38.6-kilodalton reductase (MMOR), and a 15.9-kilodalton regulatory protein (MMOB), required to couple electron consumption with substrate hydroxylation at the catalytic diiron centre of MMOH. Until now, the role of MMOB has remained ambiguous owing to a lack of atomic-level information about the MMOH-MMOB (hereafter termed H-B) complex. Here we remedy this deficiency by providing a crystal structure of H-B, which reveals the manner by which MMOB controls the conformation of residues in MMOH crucial for substrate access to the active site. MMOB docks at the α(2)β(2) interface of α(2)β(2)γ(2) MMOH, and triggers simultaneous conformational changes in the α-subunit that modulate oxygen and methane access as well as proton delivery to the diiron centre. Without such careful control by MMOB of these substrate routes to the diiron active site, the enzyme operates as an NADH oxidase rather than a monooxygenase. Biological catalysis involving small substrates is often accomplished in nature by large proteins and protein complexes. The structure presented in this work provides an elegant example of this principle.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Organizational Affiliation: