Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
Potter, J.A., Owsianka, A.M., Jeffery, N., Matthews, D.J., Keck, Z.Y., Lau, P., Foung, S.K., Taylor, G.L., Patel, A.H.(2012) J Virol 86: 12923-12932
- PubMed: 22993159
- DOI: https://doi.org/10.1128/JVI.02052-12
- Primary Citation of Related Structures:
4GAG, 4GAJ, 4GAY - PubMed Abstract:
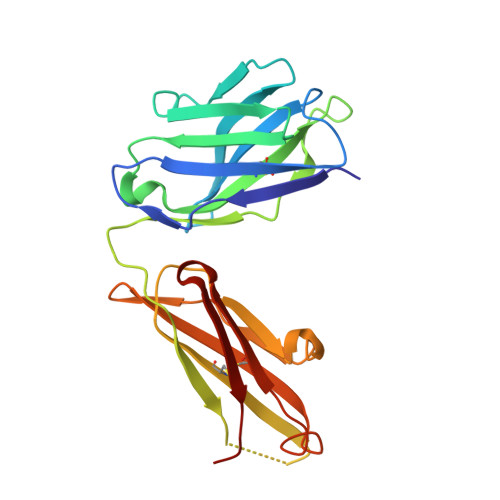
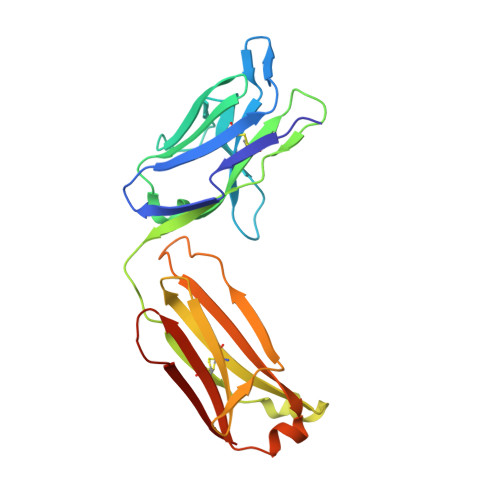
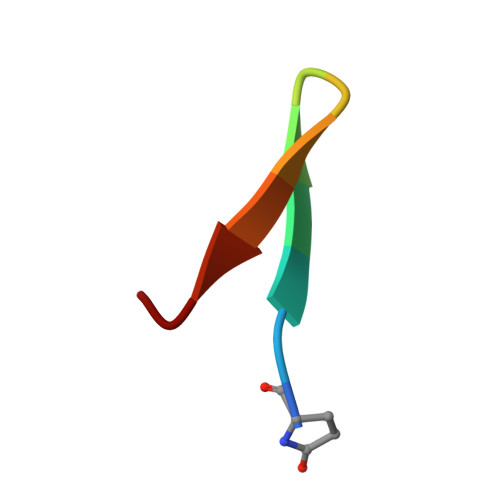
The E2 envelope glycoprotein of hepatitis C virus (HCV) binds to the host entry factor CD81 and is the principal target for neutralizing antibodies (NAbs). Most NAbs recognize hypervariable region 1 on E2, which undergoes frequent mutation, thereby allowing the virus to evade neutralization. Consequently, there is great interest in NAbs that target conserved epitopes. One such NAb is AP33, a mouse monoclonal antibody that recognizes a conserved, linear epitope on E2 and potently neutralizes a broad range of HCV genotypes. In this study, the X-ray structure of AP33 Fab in complex with an epitope peptide spanning residues 412 to 423 of HCV E2 was determined to 1.8 Å. In the complex, the peptide adopts a β-hairpin conformation and docks into a deep binding pocket on the antibody. The major determinants of antibody recognition are E2 residues L413, N415, G418, and W420. The structure is compared to the recently described HCV1 Fab in complex with the same epitope. Interestingly, the antigen-binding sites of HCV1 and AP33 are completely different, whereas the peptide conformation is very similar in the two structures. Mutagenesis of the peptide-binding residues on AP33 confirmed that these residues are also critical for AP33 recognition of whole E2, confirming that the peptide-bound structure truly represents AP33 interaction with the intact glycoprotein. The slightly conformation-sensitive character of the AP33-E2 interaction was explored by cross-competition analysis and alanine-scanning mutagenesis. The structural details of this neutralizing epitope provide a starting point for the design of an immunogen capable of eliciting AP33-like antibodies.
- Biomedical Sciences Research Complex, University of St Andrews, St Andrews, Fife, United Kingdom.
Organizational Affiliation: