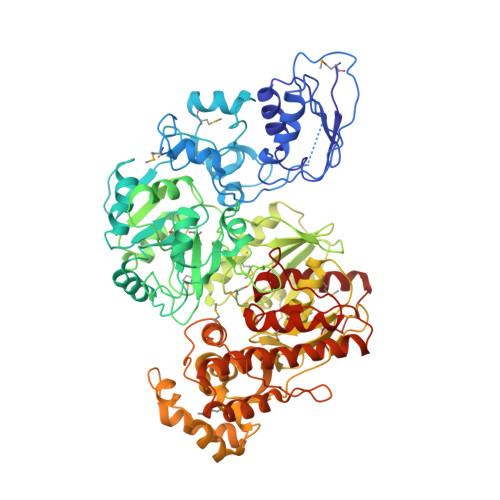
Structure of the [NiFe]-hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1.
Tominaga, T., Watanabe, S., Matsumi, R., Atomi, H., Imanaka, T., Miki, K.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1153-1157
- PubMed: 23027738
- DOI: https://doi.org/10.1107/S1744309112036421
- Primary Citation of Related Structures:
4G9I - PubMed Abstract:
HypF is involved in the biosynthesis of the CN ligand of the NiFe(CN)(2)CO centre of [NiFe]-hydrogenases. Here, the full-length structure of HypF from Thermococcus kodakarenesis is reported at 4.5 Å resolution. The N-terminal acylphosphatase-like (ACP) domain interacts with the zinc-finger domain with some flexibility in its relative position. Molecular-surface analysis shows that a deep pocket formed between the ACP and zinc-finger domains is highly conserved and has positive potential. These results suggest that the positively charged pocket identified is involved in the hydrolysis of carbamoyl phosphate and the formation of a carbamoyl intermediate.
- Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan.
Organizational Affiliation: