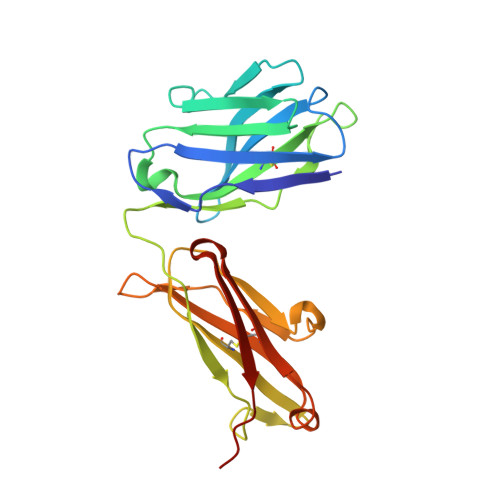
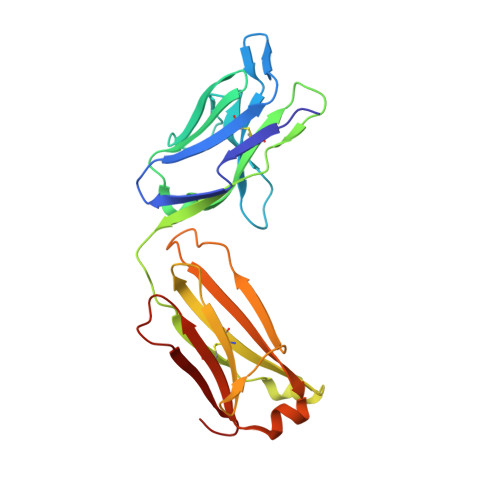
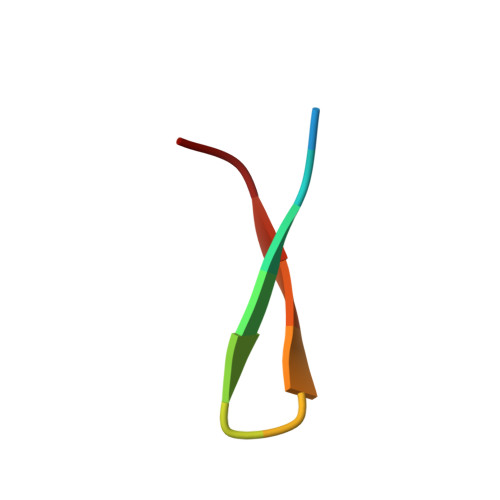Structure of Hepatitis C Virus Envelope Glycoprotein E2 Antigenic Site 412 to 423 in Complex with Antibody AP33.
Kong, L., Giang, E., Nieusma, T., Robbins, J.B., Deller, M.C., Stanfield, R.L., Wilson, I.A., Law, M.(2012) J Virol 86: 13085-13088
- PubMed: 22973046
- DOI: https://doi.org/10.1128/JVI.01939-12
- Primary Citation of Related Structures:
4G6A - PubMed Abstract:
We have determined the crystal structure of the broadly neutralizing antibody (bnAb) AP33, bound to a peptide corresponding to hepatitis C virus (HCV) E2 envelope glycoprotein antigenic site 412 to 423. Comparison with bnAb HCV1 bound to the same epitope reveals a different angle of approach to the antigen by bnAb AP33 and slight variation in its β-hairpin conformation of the epitope. These structures establish two different modes of binding to E2 that antibodies adopt to neutralize diverse HCV.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, California, USA.
Organizational Affiliation: