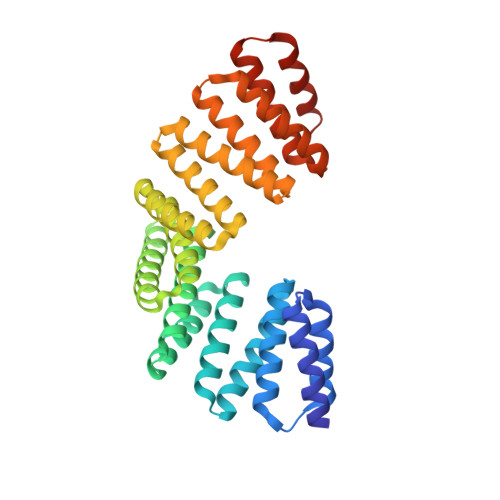
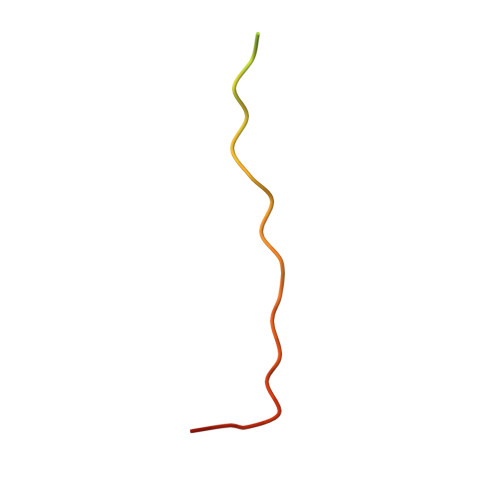
Structural and biochemical characterization of the interaction between LGN and Frmpd1
Pan, Z., Shang, Y., Jia, M., Zhang, L., Xia, C., Zhang, M., Wang, W., Wen, W.(2013) J Mol Biology 425: 1039-1049
- PubMed: 23318951
- DOI: https://doi.org/10.1016/j.jmb.2013.01.003
- Primary Citation of Related Structures:
4G2V - PubMed Abstract:
The tetratricopeptide repeat (TPR) motif-containing protein LGN binds multiple targets and regulates their subcellular localizations and functions during both asymmetric and symmetric cell divisions. Here, we characterized the interaction between LGN-TPR motifs and FERM and PDZ domain containing 1 (Frmpd1) and reported the crystal structure of the complex at 2.4Å resolution. A highly conserved fragment at the center of Frmpd1 of ~20 residues was found to be necessary and sufficient to bind to LGN-TPR. This Frmpd1 fragment forms an extended structure and runs along the concave channel of the TPR superhelix in an antiparallel manner in the complex. Structural comparisons and biochemical studies of LGN/Frmpd1 and other known LGN/target interactions demonstrate that the LGN-TPR motifs are versatile and capable of recognizing multiple targets via diverse binding modes. Nevertheless, a conserved "E/QxEx4-5E/D/Qx1-2K/R" motif in LGN/Pins (partner of inscuteable) TPR binding proteins has been identified.
- Key Laboratory of Molecular Medicine, Ministry of Education, Department of Biochemistry and Molecular Biology, and Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai 200032, P. R. China.
Organizational Affiliation: