Structural and Mechanistic Basis of Substrate Recognition by Novel Di-peptidase Dug1p From Saccromyces cerevesiae
Singh, A.K., Singh, M., Pandya, V.K., Singh, V., Mittal, M., Kumaran, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cys-Gly metallodipeptidase DUG1 | 487 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: Dug1p EC: 3.4.13 | 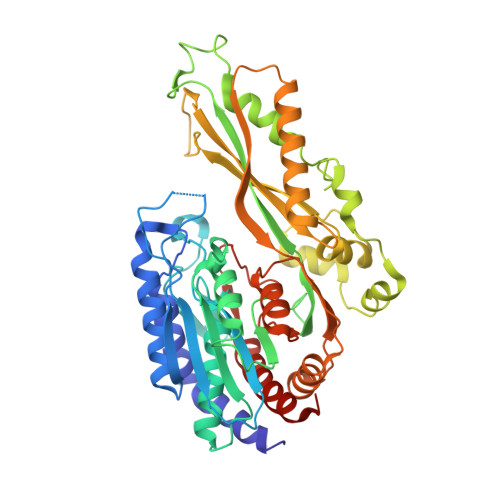 | |
UniProt | |||||
Find proteins for P43616 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P43616 Go to UniProtKB: P43616 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P43616 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CYS Query on CYS | E [auth A] | CYSTEINE C3 H7 N O2 S XUJNEKJLAYXESH-REOHCLBHSA-N |  | ||
| GLY Query on GLY | D [auth A] | GLYCINE C2 H5 N O2 DHMQDGOQFOQNFH-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | B [auth A], C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 119.132 | α = 90 |
| b = 119.132 | β = 90 |
| c = 176.302 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASES | phasing |
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |