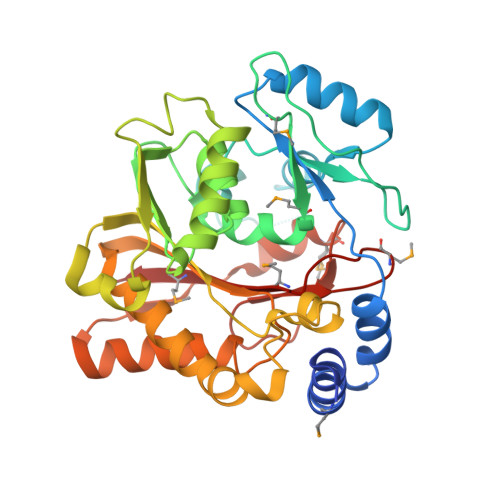
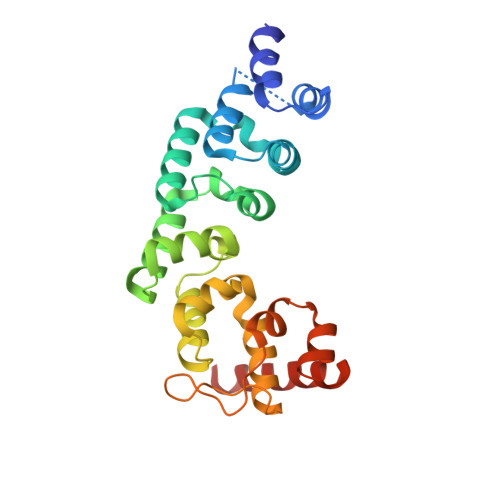
Structure of the Essential MTERF4:NSUN4 Protein Complex Reveals How an MTERF Protein Collaborates to Facilitate rRNA Modification.
Yakubovskaya, E., Guja, K.E., Mejia, E., Castano, S., Hambardjieva, E., Choi, W.S., Garcia-Diaz, M.(2012) Structure 20: 1940-1947
- PubMed: 23022348
- DOI: https://doi.org/10.1016/j.str.2012.08.027
- Primary Citation of Related Structures:
4FZV - PubMed Abstract:
MTERF4 is the first MTERF family member shown to bind RNA and plays an essential role as a regulator of ribosomal biogenesis in mammalian mitochondria. It forms a complex with the rRNA methyltransferase NSUN4 and recruits it to the large ribosomal subunit. In this article, we characterize the interaction between both proteins, demonstrate that MTERF4 strongly stimulates the specificity of NSUN4 during in vitro methylation experiments, and present the 2.0 Å resolution crystal structure of the MTERF4:NSUN4 protein complex, lacking 48 residues of the MTERF4 C-terminal acidic tail, bound to S-adenosyl-L-methionine, thus revealing the nature of the interaction between both proteins and the structural conservation of the most divergent of the human MTERF family members. Moreover, the structure suggests a model for RNA binding by the MTERF4:NSUN4 complex, providing insight into the mechanism by which an MTERF family member facilitates rRNA methylation.
- Department of Pharmacological Sciences, Stony Brook University, Stony Brook, NY 11794, USA.
Organizational Affiliation: