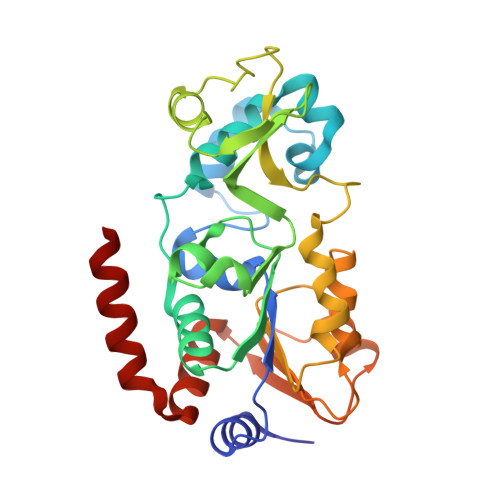
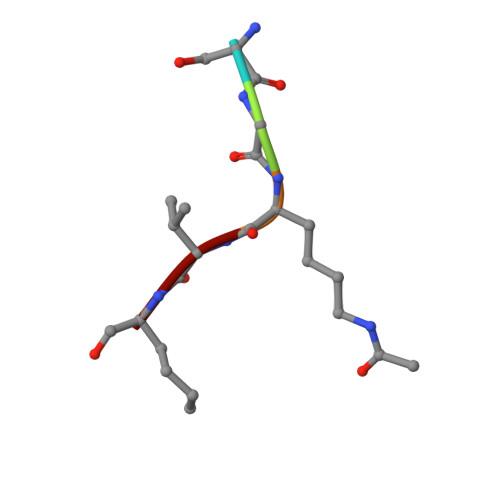
Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
Szczepankiewicz, B.G., Dai, H., Koppetsch, K.J., Qian, D., Jiang, F., Mao, C., Perni, R.B.(2012) J Org Chem 77: 7319-7329
- PubMed: 22849721
- DOI: https://doi.org/10.1021/jo301067e
- Primary Citation of Related Structures:
4FVT, 4G1C - PubMed Abstract:
Carba-NAD is a synthetic compound identical to NAD except for one substitution, where an oxygen atom adjacent to the anomeric linkage bearing nicotinamide is replaced with a methylene group. Because it is inert in nicotinamide displacement reactions, carba-NAD is an unreactive substrate analogue for NAD-consuming enzymes. SIRT3 and SIRT5 are NAD-consuming enzymes that are potential therapeutic targets for the treatment of metabolic diseases and cancers. We report an improved carba-NAD synthesis, including a pyrophosphate coupling method that proceeds in approximately 60% yield. We also disclose the X-ray crystal structures of the ternary complexes of SIRT3 and SIRT5 bound to a peptide substrate and carba-NAD. These X-ray crystal structures provide critical snapshots of the mechanism by which human sirtuins function as protein deacylation catalysts.
- Sirtris, a GSK Company, 200 Technology Square, Cambridge, Massachusetts 02139, USA. bruce.2.szczepankiewicz@gsk.com
Organizational Affiliation: