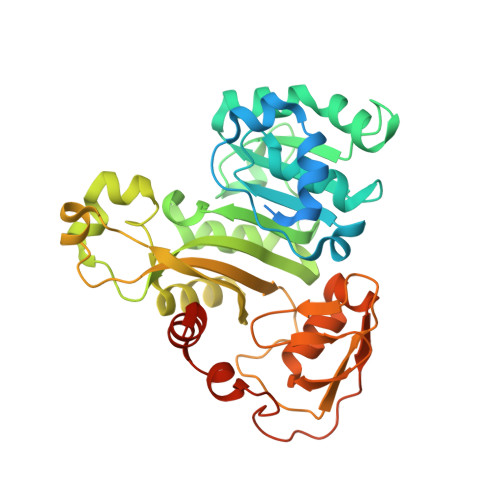
Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Ajees, A.A., Marapakala, K., Packianathan, C., Sankaran, B., Rosen, B.P.(2012) Biochemistry 51: 5476-5485
- PubMed: 22712827
- DOI: https://doi.org/10.1021/bi3004632
- Primary Citation of Related Structures:
4FR0, 4FS8, 4FSD - PubMed Abstract:
Enzymatic methylation of arsenic is a detoxification process in microorganisms but in humans may activate the metalloid to more carcinogenic species. We describe the first structure of an As(III) S-adenosylmethionine methyltransferase by X-ray crystallography that reveals a novel As(III) binding domain. The structure of the methyltransferase from the thermophilic eukaryotic alga Cyanidioschyzon merolae reveals the relationship between the arsenic and S-adenosylmethionine binding sites to a final resolution of ∼1.6 Å. As(III) binding causes little change in conformation, but binding of SAM reorients helix α4 and a loop (residues 49-80) toward the As(III) binding domain, positioning the methyl group for transfer to the metalloid. There is no evidence of a reductase domain. These results are consistent with previous suggestions that arsenic remains trivalent during the catalytic cycle. A homology model of human As(III) S-adenosylmethionine methyltransferase with the location of known polymorphisms was constructed. The structure provides insights into the mechanism of substrate binding and catalysis.
- Department of Cellular Biology and Pharmacology, Herbert Wertheim College of Medicine, Florida International University, Miami, 33199, United States.
Organizational Affiliation: