Crystal structure of transportin/FUS-NLS
Gong, W., Niu, C., Jia, M., Gao, F.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transportin-1 | 890 | Homo sapiens | Mutation(s): 0 | 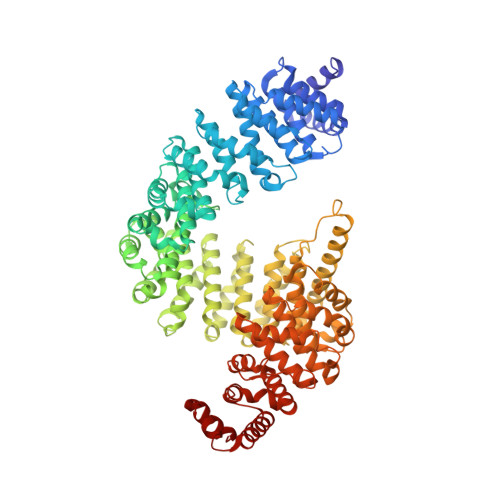 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q92973 (Homo sapiens) Explore Q92973 Go to UniProtKB: Q92973 | |||||
PHAROS: Q92973 GTEx: ENSG00000083312 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92973 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fusion (Involved in t(12;16) in malignant liposarcoma) | 37 | Homo sapiens | Mutation(s): 0 | 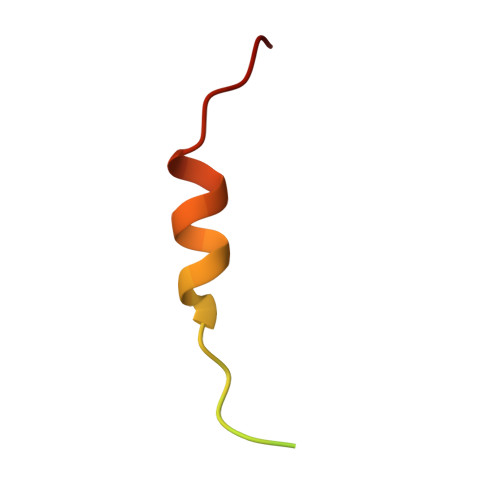 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35637 (Homo sapiens) Explore P35637 Go to UniProtKB: P35637 | |||||
PHAROS: P35637 GTEx: ENSG00000089280 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35637 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 128.811 | α = 90 |
| b = 158.292 | β = 90 |
| c = 68.37 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |