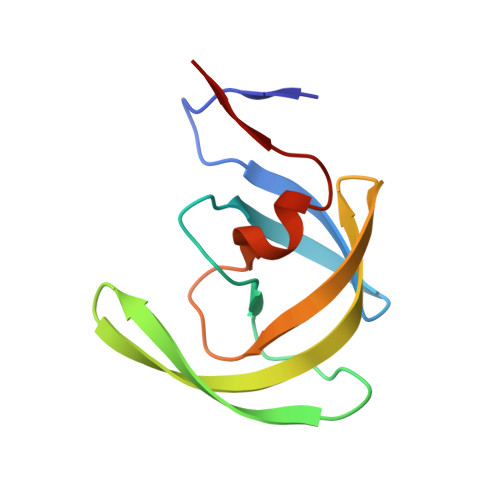
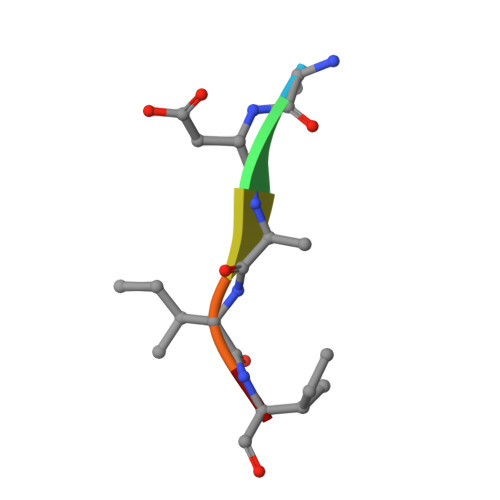
Capturing the Reaction Pathway in Near-Atomic-Resolution Crystal Structures of HIV-1 Protease.
Shen, C.H., Tie, Y., Yu, X., Wang, Y.F., Kovalevsky, A.Y., Harrison, R.W., Weber, I.T.(2012) Biochemistry 51: 7726-7732
- PubMed: 22963370
- DOI: https://doi.org/10.1021/bi3008092
- Primary Citation of Related Structures:
4FL8, 4FLG, 4FM6 - PubMed Abstract:
Snapshots of three consecutive steps in the proteolytic reaction of HIV-1 protease (PR) were obtained in crystal structures at resolutions of 1.2-1.4 Å. Structures of wild-type protease and two mutants (PR(V32I) and PR(I47V)) with V32I and I47V substitutions, which are common in drug resistance, reveal the gem-diol tetrahedral intermediate, the separating N- and C-terminal products, and the C-terminal product of an autoproteolytic peptide. These structures represent three stages in the reaction pathway and shed light on the reaction mechanism. The near-atomic-resolution geometric details include a short hydrogen bond between the intermediate and the outer carboxylate oxygen of one catalytic Asp25 that is conserved in all three structures. The two products in the complex with mutant PR(I47V) have a 2.2 Å separation of the amide and carboxyl carbon of the adjacent ends, suggesting partial cleavage prior to product release. The complex of mutant PR(V32I) with a single C-terminal product shows density for water molecules in the other half of the binding site, including a partial occupancy water molecule interacting with the product carboxylate end and the carbonyl oxygen of one conformation of Gly27, which suggests a potential role of Gly27 in recycling from the product complex to the ligand-free enzyme. These structural details at near-atomic resolution enhance our understanding of the reaction pathway and will assist in the design of mechanism-based inhibitors as antiviral agents.
- Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, Georgia 30303, United States.
Organizational Affiliation: