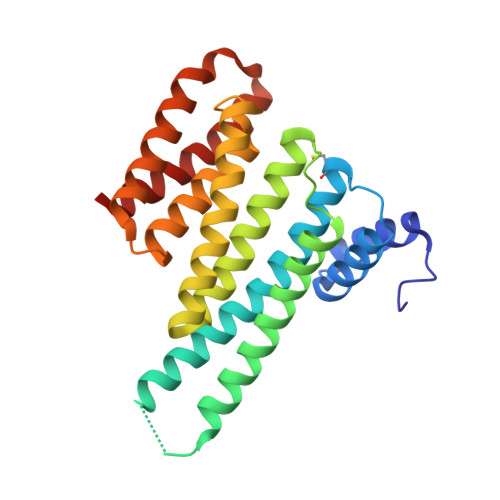
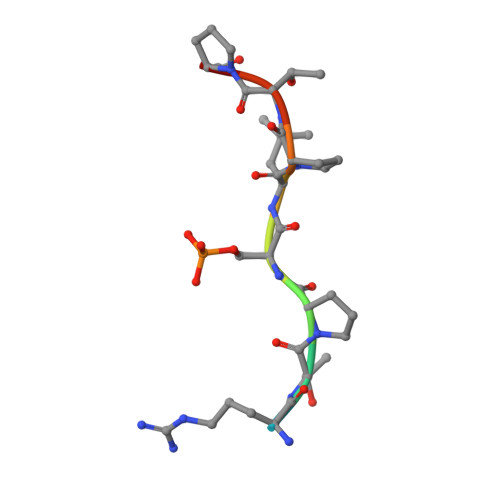
Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau.
Joo, Y., Schumacher, B., Landrieu, I., Bartel, M., Smet-Nocca, C., Jang, A., Choi, H.S., Jeon, N.L., Chang, K.A., Kim, H.S., Ottmann, C., Suh, Y.H.(2015) FASEB J 29: 4133-4144
- PubMed: 26103986
- DOI: https://doi.org/10.1096/fj.14-265009
- Primary Citation of Related Structures:
4FL5, 5BTV - PubMed Abstract:
14-3-3 proteins act as adapters that exert their function by interacting with their various protein partners. 14-3-3 proteins have been implicated in a variety of human diseases including neurodegenerative diseases. 14-3-3 proteins have recently been reported to be abundant in the neurofibrillary tangles (NFTs) observed inside the neurons of brains affected by Alzheimer's disease (AD). These NFTs are mainly constituted of phosphorylated Tau protein, a microtubule-associated protein known to bind 14-3-3. Despite this indication of 14-3-3 protein involvement in the AD pathogenesis, the role of 14-3-3 in the Tauopathy remains to be clarified. In the present study, we shed light on the role of 14-3-3 proteins in the molecular pathways leading to Tauopathies. Overexpression of the 14-3-3σ isoform resulted in a disruption of the tubulin cytoskeleton and prevented neuritic outgrowth in neurons. NMR studies validated the phosphorylated residues pSer214 and pSer324 in Tau as the 2 primary sites for 14-3-3 binding, with the crystal structure of 14-3-3σ in complex with Tau-pSer214 and Tau-pSer324 revealing the molecular details of the interaction. These data suggest a rationale for a possible pharmacologic intervention of the Tau/14-3-3 interaction.
- *Department of Pharmacology, College of Medicine and Neuroscience Research Institute, Medical Research Council, and School of Mechanical and Aerospace Engineering, Seoul National University, Seoul, Republic of Korea; Department of Pharmacology, College of Medicine, Gachon University, Incheon, Republic of Korea; Chemical Genomics Centre of the Max Planck Society, Dortmund, Germany; Unité Mixte de Recherche 8576, Centre National de la Recherche Scientifique-University of Lille, Villeneuve d'Ascq, France; Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, Eindhoven, The Netherlands; and Korea Brain Research Institute, Daegu, Republic of Korea.
Organizational Affiliation: