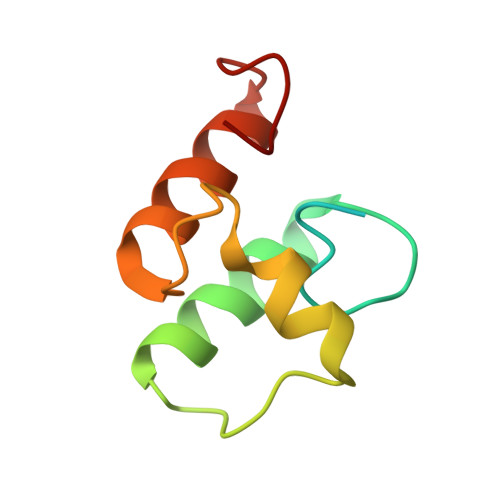
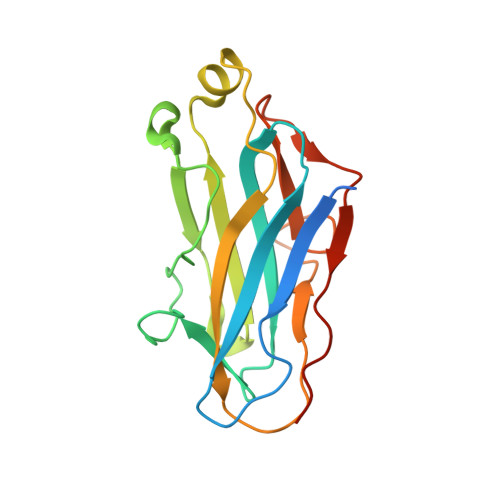
Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
Currie, M.A., Adams, J.J., Faucher, F., Bayer, E.A., Jia, Z., Smith, S.P.(2012) J Biological Chem 287: 26953-26961
- PubMed: 22707718
- DOI: https://doi.org/10.1074/jbc.M112.343897
- Primary Citation of Related Structures:
4FL4 - PubMed Abstract:
Cellulosomes are multienzyme complexes responsible for efficient degradation of plant cell wall polysaccharides. The nonenzymatic scaffoldin subunit provides a platform for cellulolytic enzyme binding that enhances the overall activity of the bound enzymes. Understanding the unique quaternary structural elements responsible for the enzymatic synergy of the cellulosome is hindered by the large size and inherent flexibility of these multiprotein complexes. Herein, we have used x-ray crystallography and small angle x-ray scattering to structurally characterize a ternary protein complex from the Clostridium thermocellum cellulosome that comprises a C-terminal trimodular fragment of the CipA scaffoldin bound to the SdbA type II cohesin module and the type I dockerin module from the Cel9D glycoside hydrolase. This complex represents the largest fragment of the cellulosome solved by x-ray crystallography to date and reveals two rigid domains formed by the type I cohesin·dockerin complex and by the X module-type II cohesin·dockerin complex, which are separated by a 13-residue linker in an extended conformation. The type I dockerin modules of the four structural models found in the asymmetric unit are in an alternate orientation to that previously observed that provides further direct support for the dual mode of binding. Conserved intermolecular contacts between symmetry-related complexes were also observed and may play a role in higher order cellulosome structure. SAXS analysis of the ternary complex revealed that the 13-residue intermodular linker of the scaffoldin subunit is highly dynamic in solution. These studies provide fundamental insights into modular positioning, linker flexibility, and higher order organization of the cellulosome.
- Department of Biomedical and Molecular Sciences, University, Kingston, Ontario K7L 3N6, Canada.
Organizational Affiliation: