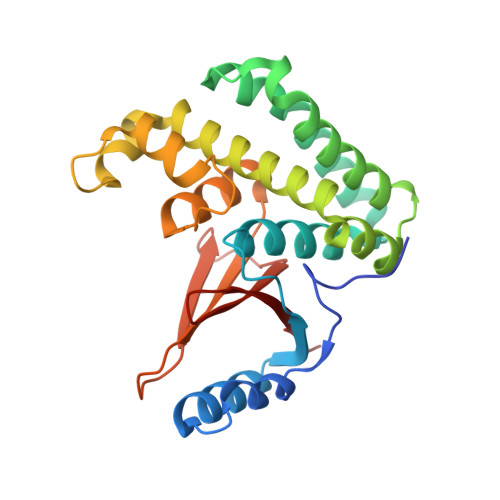
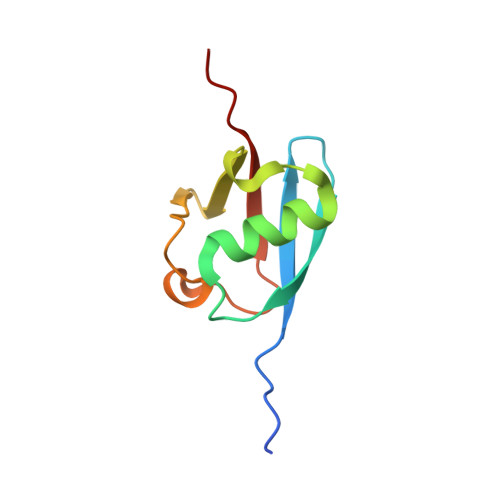
The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages.
Altun, M., Walter, T.S., Kramer, H.B., Herr, P., Iphofer, A., Bostrom, J., David, Y., Komsany, A., Ternette, N., Navon, A., Stuart, D.I., Ren, J., Kessler, B.M.(2015) PLoS One 10: e0115344-e0115344
- PubMed: 25590432
- DOI: https://doi.org/10.1371/journal.pone.0115344
- Primary Citation of Related Structures:
4FJV - PubMed Abstract:
Ovarian tumor domain containing proteases cleave ubiquitin (Ub) and ubiquitin-like polypeptides from proteins. Here we report the crystal structure of human otubain 2 (OTUB2) in complex with a ubiquitin-based covalent inhibitor, Ub-Br2. The ubiquitin binding mode is oriented differently to how viral otubains (vOTUs) bind ubiquitin/ISG15, and more similar to yeast and mammalian OTUs. In contrast to OTUB1 which has exclusive specificity towards Lys48 poly-ubiquitin chains, OTUB2 cleaves different poly-Ub linked chains. N-terminal tail swapping experiments between OTUB1 and OTUB2 revealed how the N-terminal structural motifs in OTUB1 contribute to modulating enzyme activity and Ub-chain selectivity, a trait not observed in OTUB2, supporting the notion that OTUB2 may affect a different spectrum of substrates in Ub-dependent pathways.
- Target Discovery Institute, Nuffield Department of Medicine, Roosevelt Drive, University of Oxford, Oxford, OX3 7FZ, United Kingdom; Science for Life Laboratory, Division of Translational Medicine and Chemical Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, SE-171 21 Stockholm, Sweden.
Organizational Affiliation: