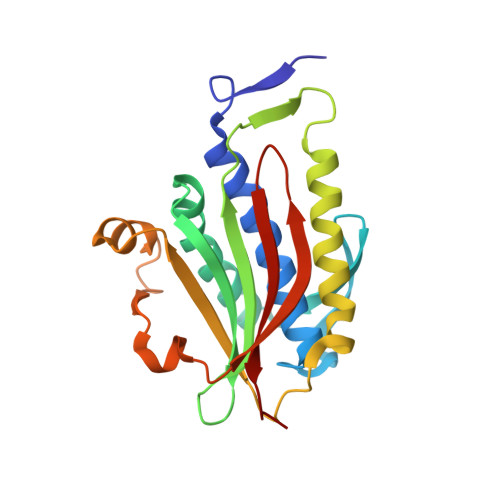
Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
Wojtaszek, J., Lee, C.J., D'Souza, S., Minesinger, B., Kim, H., D'Andrea, A.D., Walker, G.C., Zhou, P.(2012) J Biological Chem 287: 33836-33846
- PubMed: 22859295
- DOI: https://doi.org/10.1074/jbc.M112.394841
- Primary Citation of Related Structures:
4FJO - PubMed Abstract:
DNA synthesis across lesions during genomic replication requires concerted actions of specialized DNA polymerases in a potentially mutagenic process known as translesion synthesis. Current models suggest that translesion synthesis in mammalian cells is achieved in two sequential steps, with a Y-family DNA polymerase (κ, η, ι, or Rev1) inserting a nucleotide opposite the lesion and with the heterodimeric B-family polymerase ζ, consisting of the catalytic Rev3 subunit and the accessory Rev7 subunit, replacing the insertion polymerase to carry out primer extension past the lesion. Effective translesion synthesis in vertebrates requires the scaffolding function of the C-terminal domain (CTD) of Rev1 that interacts with the Rev1-interacting region of polymerases κ, η, and ι and with the Rev7 subunit of polymerase ζ. We report the purification and structure determination of a quaternary translesion polymerase complex consisting of the Rev1 CTD, the heterodimeric Pol ζ complex, and the Pol κ Rev1-interacting region. Yeast two-hybrid assays were employed to identify important interface residues of the translesion polymerase complex. The structural elucidation of such a quaternary translesion polymerase complex encompassing both insertion and extension polymerases bridged by the Rev1 CTD provides the first molecular explanation of the essential scaffolding function of Rev1 and highlights the Rev1 CTD as a promising target for developing novel cancer therapeutics to suppress translesion synthesis. Our studies support the notion that vertebrate insertion and extension polymerases could structurally cooperate within a megatranslesion polymerase complex (translesionsome) nucleated by Rev1 to achieve efficient lesion bypass without incurring an additional switching mechanism.
- Department of Biochemistry, Duke University, Medical Center, Durham, North Carolina 27710, USA.
Organizational Affiliation: