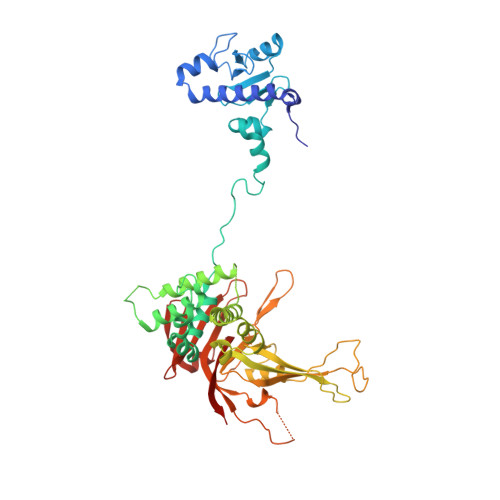
A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Samara, N.L., Ringel, A.E., Wolberger, C.(2012) Structure 20: 1414-1424
- PubMed: 22771212
- DOI: https://doi.org/10.1016/j.str.2012.05.015
- Primary Citation of Related Structures:
4FIP, 4FJC, 4FK5 - PubMed Abstract:
The deubiquitinating module (DUBm) of the SAGA coactivator contains the Ubp8 isopeptidase, Sgf11, Sus1, and Sgf73, which form a highly interconnected complex. Although Ubp8 contains a canonical USP catalytic domain, it is only active when in complex with the other DUBm subunits. The Sgf11 zinc finger (Sgf11-ZnF) binds near the Ubp8 active site and is essential for full activity, suggesting that the Sgf11-ZnF helps maintain the active conformation of Ubp8. We report structural and solution studies showing that deletion of the Sgf11-ZnF destabilizes incorporation of Ubp8 within the DUBm, giving rise to domain swapping with a second complex and misaligning active site residues. Activating mutations in Ubp8 that partially restore activity in the absence of the Sgf11-ZnF promote the monomeric form of the DUBm. Our data suggest an unexpected role for Sgf11 in compensating for the absence of structural features that maintain the active conformation of Ubp8.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, 725 N. Wolfe Street, Baltimore, MD 21205-2185, USA.
Organizational Affiliation: