Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS).
Zhang, Z.C., Chook, Y.M.(2012) Proc Natl Acad Sci U S A 109: 12017-12021
- PubMed: 22778397
- DOI: https://doi.org/10.1073/pnas.1207247109
- Primary Citation of Related Structures:
4FDD - PubMed Abstract:
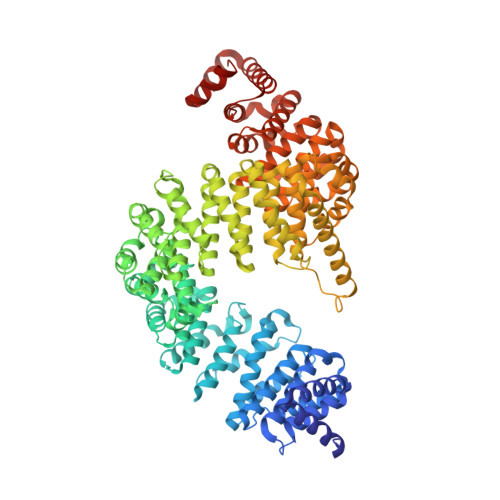
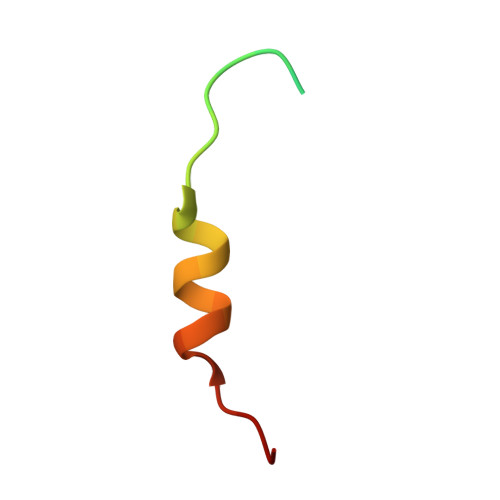
Mutations in the proline/tyrosine-nuclear localization signal (PY-NLS) of the Fused in Sarcoma protein (FUS) cause amyotrophic lateral sclerosis (ALS). Here we report the crystal structure of the FUS PY-NLS bound to its nuclear import receptor Karyopherinβ2 (Kapβ2; also known as Transportin). The FUS PY-NLS occupies the structurally invariant C-terminal arch of Kapβ2, tracing a path similar to that of other characterized PY-NLSs. Unlike other PY-NLSs, which generally bind Kapβ2 in fully extended conformations, the FUS peptide is atypical as its central portion forms a 2.5-turn α-helix. The Kapβ2-binding epitopes of the FUS PY-NLS consist of an N-terminal PGKM hydrophobic motif, a central arginine-rich α-helix, and a C-terminal PY motif. ALS mutations are found almost exclusively within these epitopes. Each ALS mutation site makes multiple contacts with Kapβ2 and mutations of these residues decrease binding affinities for Kapβ2 (K(D) for wild-type FUS PY-NLS is 9.5 nM) up to ninefold. Thermodynamic analyses of ALS mutations in the FUS PY-NLS show that the weakening of FUS-Kapβ2 binding affinity, the degree of cytoplasmic mislocalization, and ALS disease severity are correlated.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: